Tutorial 7.6b - Factorial ANOVA (Bayesian)
12 Jan 2018
Overview
Factorial designs are an extension of single factor ANOVA designs in which additional factors are added such that each level of one factor is applied to all levels of the other factor(s) and these combinations are replicated.
For example, we might design an experiment in which the effects of temperature (high vs low) and fertilizer (added vs not added) on the growth rate of seedlings are investigated by growing seedlings under the different temperature and fertilizer combinations.
The following diagram depicts a very simple factorial design in which there are two factors (Shape and Color) each with two levels (Shape: square and circle; Color: blue and white) and each combination has 3 replicates.
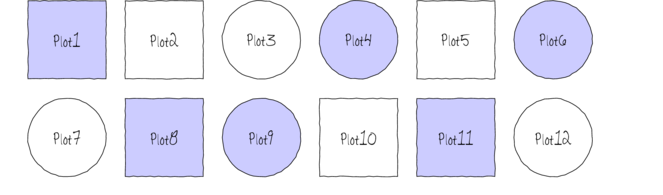
In addition to investigating the impacts of the main factors, factorial designs allow us to investigate whether the effects of one factor are consistent across levels of another factor. For example, is the effect of temperature on growth rate the same for both fertilized and unfertilized seedlings and similarly, does the impact of fertilizer treatment depend on the temperature under which the seedlings are grown?
Arguably, these interactions give more sophisticated insights into the dynamics of the system we are investigating. Hence, we could add additional main effects, such as soil pH, amount of water etc along with all the two way (temp:fert, temp:pH, temp:water, etc), three-way (temp:fert:pH, temp:pH:water), four-way etc interactions in order to explore how these various factors interact with one another to effect the response.
However, the more interactions, the more complex the model becomes to specify, compute and interpret - not to mention the rate at which the number of required observations increases.
To appreciate the interpretation of interactions, consider the following figure that depicts fictitious two factor (temperature and fertilizer) designs.
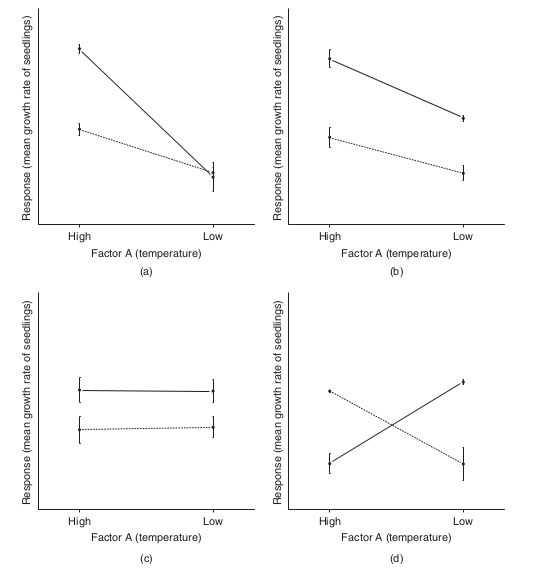
It is clear from the top-right figure that whether or not there is an observed effect of adding fertilizer or not depends on whether we are focused on seedlings growth under high or low temperatures. Fertilizer is only important for seedlings grown under high temperatures. In this case, it is not possible to simply state that there is an effect of fertilizer as it depends on the level of temperature. Similarly, the magnitude of the effect of temperature depends on whether fertilizer has been added or not.
Such interactions are represented by plots in which lines either intersect or converge. The top-right and bottom-left figures both depict parallel lines which are indicative of no interaction. That is, the effects of temperature are similar for both fertilizer added and controls and vice verse. Whilst the former displays an effect of both fertilizer and temperature, the latter only fertilizer is important.
Finally, the bottom-right figure represents a strong interaction that would mask the main effects of temperature and fertilizer (since the nature of the effect of temperature is very different for the different fertilizer treatments and visa verse).
In a frequentist framework, factorial designs can consist:
- entirely of crossed fixed factors (Model I ANOVA - most common) in which conclusions are restricted to the specific combinations of levels selected for the experiment,
- entirely of crossed random factors (Model II ANOVA) or
- a mixture of crossed fixed and random factors (Model III ANOVA).
The tutorial on frequentist factorial ANOVA described procedures used to further investigate models in the presence of significant interactions as well as the complications that arise with linear models fitted to unbalanced designs. Again, these issues largely evaporate in a Bayesian framework. Consequently, we will not really dwell on these complications in this tutorial. At most, we will model some unbalanced data, yet it should be noted that we will not need to make any special adjustments in order to do so.
Linear model
The generic linear model is presented here purely for revisory purposes. If, it is unfamiliar to you or you are unsure about what parameters are to be estimated and tested, you are strongly advised to review the the tutorial on frequentist factorial ANOVA
The linear models for two and three factor design are: $$y_{ijk}=\mu+\alpha_i + \beta_{j} + (\alpha\beta)_{ij} + \varepsilon_{ijk}$$ $$y_{ijkl}=\mu+\alpha_i + \beta_{j} + \gamma_{k} + (\alpha\beta)_{ij} + (\alpha\gamma)_{ik} + (\beta\gamma)_{jk} + (\alpha\beta\gamma)_{ijk} + \varepsilon_{ijkl}$$ where $\mu$ is the overall mean, $\alpha$ is the effect of Factor A, $\beta$ is the effect of Factor B, $\gamma$ is the effect of Factor C and $\varepsilon$ is the random unexplained or residual component.
Scenario and Data
Imagine we has designed an experiment in which we had measured the response ($y$) under a combination of two different potential influences (Factor A: levels $a1$ and $a2$; and Factor B: levels $b1$, $b2$ and $b3$), each combination replicated 10 times ($n=10$). As this section is mainly about the generation of artificial data (and not specifically about what to do with the data), understanding the actual details are optional and can be safely skipped. Consequently, I have folded (toggled) this section away.
- the sample size per treatment=10
- factor A with 2 levels
- factor B with 3 levels
- the 6 effects parameters are 40, 15, 5, 0, -15,10 ($\mu_{a1b1}=40$, $\mu_{a2b1}=40+15=55$, $\mu_{a1b2}=40+5=45$, $\mu_{a1b3}=40+0=40$, $\mu_{a2b2}=40+15+5-15=45$ and $\mu_{a2b3}=40+15+0+10=65$)
- the data are drawn from normal distributions with a mean of 0 and standard deviation of 3 ($\sigma^2=9$)
set.seed(1) nA <- 2 #number of levels of A nB <- 3 #number of levels of B nsample <- 10 #number of reps in each A <- gl(nA, 1, nA, lab = paste("a", 1:nA, sep = "")) B <- gl(nB, 1, nB, lab = paste("b", 1:nB, sep = "")) data <- expand.grid(A = A, B = B, n = 1:nsample) X <- model.matrix(~A * B, data = data) eff <- c(40, 15, 5, 0, -15, 10) sigma <- 3 #residual standard deviation n <- nrow(data) eps <- rnorm(n, 0, sigma) #residuals data$y <- as.numeric(X %*% eff + eps) head(data) #print out the first six rows of the data set
A B n y 1 a1 b1 1 38.12064 2 a2 b1 1 55.55093 3 a1 b2 1 42.49311 4 a2 b2 1 49.78584 5 a1 b3 1 40.98852 6 a2 b3 1 62.53859
with(data, interaction.plot(A, B, y))
## ALTERNATIVELY, we could supply the population means and get the effect parameters from these. To ## correspond to the model matrix, enter the population means in the order of: a1b1, a2b1, a1b1, ## a2b2,a1b3,a2b3 pop.means <- as.matrix(c(40, 55, 45, 45, 40, 65), byrow = F) ## Generate a minimum model matrix for the effects XX <- model.matrix(~A * B, expand.grid(A = factor(1:2), B = factor(1:3))) ## Use the solve() function to solve what are effectively simultaneous equations (eff <- as.vector(solve(XX, pop.means)))
[1] 40 15 5 0 -15 10
data$y <- as.numeric(X %*% eff + eps)
With these sort of data, we are primarily interested in investigating whether there is a relationship between the continuous response variable and the treatment type. Does treatment type effect the response.
Assumptions
- All of the observations are independent - this must be addressed at the design and collection stages. Importantly, to be considered independent replicates, the replicates must be made at the same scale at which the treatment is applied. For example, if the experiment involves subjecting organisms housed in tanks to different water temperatures, then the unit of replication is the individual tanks not the individual organisms in the tanks. The individuals in a tank are strictly not independent with respect to the treatment.
- The response variable (and thus the residuals) should be normally distributed for each sampled populations (combination of factors). Boxplots of each treatment combination are useful for diagnosing major issues with normality.
- The response variable should be equally varied (variance should not be related to mean as these are supposed to be estimated separately) for each combination of treatments. Again, boxplots are useful.
Exploratory data analysis
Normality and Homogeneity of variance
boxplot(y ~ A * B, data)

# OR via ggplot2 library(ggplot2) ggplot(data, aes(y = y, x = A, fill = B)) + geom_boxplot()

Conclusions:
- there is no evidence that the response variable is consistently non-normal across all populations - each boxplot is approximately symmetrical
- there is no evidence that variance (as estimated by the height of the boxplots) differs between the five populations. . More importantly, there is no evidence of a relationship between mean and variance - the height of boxplots does not increase with increasing position along the y-axis. Hence it there is no evidence of non-homogeneity
- transform the scale of the response variables (to address normality etc). Note transformations should be applied to the entire response variable (not just those populations that are skewed).
Model fitting or statistical analysis
It is possible to model in all sorts of specific comparisons (contrasts) into a JAGS or STAN model statement. Likewise, it is possible to define specific main effects type tests within the model statement. However, for consistency across each of the routines, we will just define the minimum required model and perform all other posterior derivatives (such as main effects tests and contrasts) from the MCMC samples using R. This way, the techniques can be applied no mater which of the Bayesian modelling routines (JAGS, STAN, MCMCpack etc) were used.
The observed response ($y_i$) are assumed to be drawn from a normal distribution with a given mean ($\mu$) and standard deviation ($\sigma$). The expected values ($\mu$) are themselves determined by the linear predictor ($\mathbf{X}\boldsymbol{\beta}$). In this case, $\boldsymbol{\beta}$ represents the vector of $\beta$'s - the intercept associated with the first combination of groups, as well as the (effects) differences between this intercept and each other group. $\mathbf{X}$ is the model matrix.
MCMC sampling requires priors on all parameters. We will employ weakly informative priors. Specifying 'uninformative' priors is always a bit of a balancing act. If the priors are too vague (wide) the MCMC sampler can wander off into nonscence areas of likelihood rather than concentrate around areas of highest likelihood (desired when wanting the outcomes to be largely driven by the data). On the other hand, if the priors are too strong, they may have an influence on the parameters. In such a simple model, this balance is very forgiving - it is for more complex models that prior choice becomes more important.
For this simple model, we will go with zero-centered Gaussian (normal) priors with relatively large standard deviations (100) for both the intercept and the treatment effect and a wide half-cauchy (scale=5) for the standard deviation. $$ \begin{align} y_{ij} &\sim{} N(\mu_{ij}, \sigma)\\ \mu_{ij} &= \beta_0 + \mathbf{X}\boldsymbol{\beta}\\[1em] \beta_0 &\sim{} N(0,100)\\ \beta &\sim{} N(0,10)\\ \sigma &\sim{} cauchy(0,5)\\ \end{align} $$ Exploratory data analysis suggests that the intercept and effects could be drawn from similar distributions (with mean in the 10's and variances in the 100's). Whilst we might therefore be tempted to provide different priors for the intercept, compared to the effects, for a simple model such as this, it is unlikely to be necessary. However, for more complex models, where prior specification becomes more critical, separate priors would probably be necessary.
library(MCMCpack) data.mcmcpack <- MCMCregress(y ~ A * B, data = data)
Define the model
modelString = " model { #Likelihood for (i in 1:n) { y[i]~dnorm(mean[i],tau) mean[i] <- inprod(beta[],X[i,]) } #Priors for (i in 1:ngroups) { beta[i] ~ dnorm(0, 1.0E-6) } sigma ~ dunif(0, 100) tau <- 1 / (sigma * sigma) } "
Define the data
Arrange the data as a list (as required by BUGS). As input, JAGS will need to be supplied with:
- the response variable (y)
- the predictor model matrix (X)
- the total number of observed items (n)
- the number of predictor terms (nX)
X <- model.matrix(~A * B, data) data.list <- with(data, list(y = y, X = X, n = nrow(data), ngroups = ncol(X)))
Define the MCMC chain parameters
Next we should define the behavioural parameters of the MCMC sampling chains. Include the following:
- the nodes (estimated parameters) to monitor (return samples for)
- the number of MCMC chains (3)
- the number of burnin steps (1000)
- the thinning factor (10)
- the number of MCMC iterations - determined by the number of samples to save, the rate of thinning and the number of chains
params <- c("beta", "sigma") nChains = 3 burnInSteps = 3000 thinSteps = 10 numSavedSteps = 15000 #across all chains nIter = ceiling(burnInSteps + (numSavedSteps * thinSteps)/nChains) nIter
[1] 53000
Fit the model
Now run the JAGS code via the R2jags interface. Note that the first time jags is run after the R2jags package is loaded, it is often necessary to run any kind of randomization function just to initiate the .Random.seed variable.
library(R2jags)
data.r2jags <- jags(data = data.list, inits = NULL, parameters.to.save = params, model.file = textConnection(modelString), n.chains = nChains, n.iter = nIter, n.burnin = burnInSteps, n.thin = thinSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph information: Observed stochastic nodes: 60 Unobserved stochastic nodes: 7 Total graph size: 514 Initializing model
print(data.r2jags)
Inference for Bugs model at "5", fit using jags,
3 chains, each with 53000 iterations (first 3000 discarded), n.thin = 10
n.sims = 15000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta[1] 41.094 0.844 39.441 40.530 41.095 41.652 42.743 1.001 15000
beta[2] 14.647 1.194 12.274 13.867 14.652 15.442 16.982 1.001 15000
beta[3] 4.652 1.193 2.295 3.852 4.654 5.441 6.977 1.001 15000
beta[4] -0.746 1.190 -3.126 -1.515 -0.736 0.046 1.597 1.001 15000
beta[5] -15.712 1.695 -19.045 -16.834 -15.721 -14.601 -12.325 1.001 15000
beta[6] 9.336 1.673 6.039 8.232 9.335 10.434 12.585 1.001 15000
sigma 2.662 0.261 2.210 2.478 2.639 2.826 3.227 1.001 15000
deviance 286.111 4.072 280.390 283.115 285.406 288.308 295.927 1.001 15000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 8.3 and DIC = 294.4
DIC is an estimate of expected predictive error (lower deviance is better).
data.mcmc.list <- as.mcmc(data.r2jags)
Whilst Gibbs sampling provides an elegantly simple MCMC sampling routine, very complex hierarchical models can take enormous numbers of iterations (often prohibitory large) to converge on a stable posterior distribution.
To address this, Andrew Gelman (and other collaborators) have implemented a variation on Hamiltonian Monte Carlo (HMC: a sampler that selects subsequent samples in a way that reduces the correlation between samples, thereby speeding up convergence) called the No-U-Turn (NUTS) sampler. All of these developments are brought together into a tool called Stan ("Sampling Through Adaptive Neighborhoods").
By design (to appeal to the vast BUGS users), Stan models are defined in a manner reminiscent of BUGS. Stan first converts these models into C++ code which is then compiled to allow very rapid computation.
Consistent with the use of C++, the model must be accompanied by variable declarations for all inputs and parameters.
One important difference between Stan and JAGS is that whereas BUGS (and thus JAGS) use precision rather than variance, Stan uses variance.
Stan itself is a stand-alone command line application. However, conveniently, the authors of Stan have also developed an R interface to Stan called Rstan which can be used much like R2jags.
Model matrix formulation
The minimum model in Stan required to fit the above simple regression follows. Note the following modifications from the model defined in JAGS:- the normal distribution is defined by variance rather than precision
- rather than using a uniform prior for sigma, I am using a half-Cauchy
We now translate the likelihood model into STAN code.
$$\begin{align}
y_{ij}&\sim{}N(\mu_{ij}, \sigma)\\
\mu_{ij} &= \mathbf{X}\boldsymbol{\beta}\\
\beta_0&\sim{}N(0,100)\\
\beta&\sim{}N(0,10)\\
\sigma&\sim{}Cauchy(0,5)\\
\end{align}
$$
Define the model
modelString = " data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } transformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,100); sigma~cauchy(0,5); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } "
Define the data
Arrange the data as a list (as required by BUGS). As input, JAGS will need to be supplied with:
- the response variable (y)
- the predictor model matrix (X)
- the total number of observed items (n)
- the number of predictor terms (nX)
Xmat <- model.matrix(~A * B, data) data.list <- with(data, list(y = y, X = Xmat, nX = ncol(Xmat), n = nrow(data)))
Fit the model
Now run the JAGS code via the R2jags interface. Note that the first time jags is run after the R2jags package is loaded, it is often necessary to run any kind of randomization function just to initiate the .Random.seed variable.
## load the rstan package library(rstan)
data.rstan <- stan(data = data.list, model_code = modelString, chains = 3, iter = 2000, warmup = 500, thin = 3)
In file included from /usr/local/lib/R/site-library/BH/include/boost/config.hpp:39:0,
from /usr/local/lib/R/site-library/BH/include/boost/math/tools/config.hpp:13,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/var.hpp:7,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/gevv_vvv_vari.hpp:5,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core.hpp:12,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/mat.hpp:4,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math.hpp:4,
from /usr/local/lib/R/site-library/StanHeaders/include/src/stan/model/model_header.hpp:4,
from file1cc41099a43b.cpp:8:
/usr/local/lib/R/site-library/BH/include/boost/config/compiler/gcc.hpp:186:0: warning: "BOOST_NO_CXX11_RVALUE_REFERENCES" redefined
# define BOOST_NO_CXX11_RVALUE_REFERENCES
^
<command-line>:0:0: note: this is the location of the previous definition
SAMPLING FOR MODEL '3d2414c9dcf4b5e12be870eadd2c894a' NOW (CHAIN 1).
Gradient evaluation took 1.7e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 501 / 2000 [ 25%] (Sampling)
Iteration: 700 / 2000 [ 35%] (Sampling)
Iteration: 900 / 2000 [ 45%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
Iteration: 1300 / 2000 [ 65%] (Sampling)
Iteration: 1500 / 2000 [ 75%] (Sampling)
Iteration: 1700 / 2000 [ 85%] (Sampling)
Iteration: 1900 / 2000 [ 95%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.051944 seconds (Warm-up)
0.117211 seconds (Sampling)
0.169155 seconds (Total)
SAMPLING FOR MODEL '3d2414c9dcf4b5e12be870eadd2c894a' NOW (CHAIN 2).
Gradient evaluation took 8e-06 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.08 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 501 / 2000 [ 25%] (Sampling)
Iteration: 700 / 2000 [ 35%] (Sampling)
Iteration: 900 / 2000 [ 45%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
Iteration: 1300 / 2000 [ 65%] (Sampling)
Iteration: 1500 / 2000 [ 75%] (Sampling)
Iteration: 1700 / 2000 [ 85%] (Sampling)
Iteration: 1900 / 2000 [ 95%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.050512 seconds (Warm-up)
0.117971 seconds (Sampling)
0.168483 seconds (Total)
SAMPLING FOR MODEL '3d2414c9dcf4b5e12be870eadd2c894a' NOW (CHAIN 3).
Gradient evaluation took 9e-06 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 501 / 2000 [ 25%] (Sampling)
Iteration: 700 / 2000 [ 35%] (Sampling)
Iteration: 900 / 2000 [ 45%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
Iteration: 1300 / 2000 [ 65%] (Sampling)
Iteration: 1500 / 2000 [ 75%] (Sampling)
Iteration: 1700 / 2000 [ 85%] (Sampling)
Iteration: 1900 / 2000 [ 95%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.053997 seconds (Warm-up)
0.118393 seconds (Sampling)
0.17239 seconds (Total)
print(data.rstan, par = c("beta", "sigma"))
Inference for Stan model: 3d2414c9dcf4b5e12be870eadd2c894a.
3 chains, each with iter=2000; warmup=500; thin=3;
post-warmup draws per chain=500, total post-warmup draws=1500.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
beta[1] 41.13 0.03 0.84 39.47 40.54 41.16 41.68 42.74 1070 1
beta[2] 14.60 0.04 1.17 12.36 13.80 14.62 15.36 16.94 1026 1
beta[3] 4.60 0.04 1.20 2.28 3.81 4.59 5.40 6.89 1134 1
beta[4] -0.79 0.04 1.17 -3.07 -1.55 -0.81 -0.03 1.65 1065 1
beta[5] -15.64 0.05 1.61 -18.74 -16.74 -15.64 -14.51 -12.48 1141 1
beta[6] 9.38 0.05 1.63 6.22 8.27 9.38 10.51 12.69 1080 1
sigma 2.66 0.01 0.26 2.21 2.47 2.63 2.83 3.24 1232 1
Samples were drawn using NUTS(diag_e) at Sat Nov 25 17:19:17 2017.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
The STAN team has put together pre-compiled modules (functions) to make specifying and applying STAN models much simpler. Each function offers a consistent interface that is Also reminiscent of major frequentist linear modelling routines in R.
Whilst it is not necessary to specify priors when using rstanarm functions (as defaults will be generated), there is no guarantee that the routines for determining these defaults will persist over time. Furthermore, it is always better to define your own priors if for no other reason that it forces you to thing about what you re doing. Consistent with the pure STAN version, we will employ the following priors:
- weakly informative Gaussian prior for the intercept $\beta_0 \sim{} N(0, 100)$
- weakly informative Gaussian prior for the treatment effect $\beta_1 \sim{} N(0, 100)$
- half-cauchy prior for the variance $\sigma \sim{} Cauchy(0, 5)$
Note, I am using the refresh=0 option so as to suppress the larger regular output in the interest of keeping output to what is necessary for this tutorial. When running outside of a tutorial context, the regular verbose output is useful as it provides a way to gauge progress.
library(rstanarm) library(broom) library(coda)
data.rstanarm = stan_glm(y ~ A * B, data = data, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior_intercept = normal(0, 100), prior = normal(0, 100), prior_aux = cauchy(0, 2))
Gradient evaluation took 4.9e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.49 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.154465 seconds (Warm-up)
0.321999 seconds (Sampling)
0.476464 seconds (Total)
Gradient evaluation took 1.6e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.16 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.195459 seconds (Warm-up)
0.301423 seconds (Sampling)
0.496882 seconds (Total)
Gradient evaluation took 1.4e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.135384 seconds (Warm-up)
0.281972 seconds (Sampling)
0.417356 seconds (Total)
print(data.rstanarm)
stan_glm
family: gaussian [identity]
formula: y ~ A * B
------
Estimates:
Median MAD_SD
(Intercept) 41.1 0.9
Aa2 14.7 1.2
Bb2 4.7 1.2
Bb3 -0.7 1.2
Aa2:Bb2 -15.7 1.6
Aa2:Bb3 9.3 1.7
sigma 2.6 0.3
Sample avg. posterior predictive
distribution of y (X = xbar):
Median MAD_SD
mean_PPD 48.7 0.5
------
For info on the priors used see help('prior_summary.stanreg').
tidyMCMC(data.rstanarm, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 (Intercept) 41.0695427 0.8462585 39.407984 42.715661 2 Aa2 14.6591690 1.1981562 12.398261 17.112206 3 Bb2 4.6819518 1.1857841 2.323231 6.980180 4 Bb3 -0.7107484 1.1808212 -2.927747 1.642205 5 Aa2:Bb2 -15.7289876 1.6946307 -19.022967 -12.286452 6 Aa2:Bb3 9.2932738 1.7176328 6.132746 12.911395 7 sigma 2.6549943 0.2653598 2.169464 3.191681
The brms package serves a similar goal to the rstanarm package - to provide a simple user interface to STAN. However, unlike the rstanarm implementation, brms simply converts the formula, data, priors and family into STAN model code and data before executing stan with those elements.
Whilst it is not necessary to specify priors when using brms functions (as defaults will be generated), there is no guarantee that the routines for determining these defaults will persist over time. Furthermore, it is always better to define your own priors if for no other reason that it forces you to thing about what you are doing. Consistent with the pure STAN version, we will employ the following priors:
- weakly informative Gaussian prior for the intercept $\beta_0 \sim{} N(0, 100)$
- weakly informative Gaussian prior for the treatment effect $\beta_1 \sim{} N(0, 100)$
- half-cauchy prior for the variance $\sigma \sim{} Cauchy(0, 5)$
Note, I am using the refresh=0. option so as to suppress the larger regular output in the interest of keeping output to what is necessary for this tutorial. When running outside of a tutorial context, the regular verbose output is useful as it provides a way to gauge progress.
library(brms) library(broom) library(coda)
data.brms = brm(y ~ A * B, data = data, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior = c(prior(normal(0, 100), class = "Intercept"), prior(normal(0, 100), class = "b"), prior(cauchy(0, 5), class = "sigma")))
Gradient evaluation took 1.5e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.059325 seconds (Warm-up)
0.110061 seconds (Sampling)
0.169386 seconds (Total)
Gradient evaluation took 9e-06 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.053149 seconds (Warm-up)
0.109058 seconds (Sampling)
0.162207 seconds (Total)
Gradient evaluation took 9e-06 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.05343 seconds (Warm-up)
0.107953 seconds (Sampling)
0.161383 seconds (Total)
print(data.brms)
Family: gaussian(identity)
Formula: y ~ A * B
Data: data (Number of observations: 60)
Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2;
total post-warmup samples = 2250
ICs: LOO = NA; WAIC = NA; R2 = NA
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
Intercept 41.12 0.85 39.44 42.92 1936 1
Aa2 14.63 1.20 12.27 16.87 1538 1
Bb2 4.62 1.20 2.29 7.01 1914 1
Bb3 -0.76 1.21 -3.13 1.58 2013 1
Aa2:Bb2 -15.68 1.71 -18.92 -12.25 1696 1
Aa2:Bb3 9.31 1.72 6.00 12.66 1713 1
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
sigma 2.64 0.26 2.2 3.21 1401 1
Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
is a crude measure of effective sample size, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).
tidyMCMC(data.brms, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 b_Intercept 41.1249801 0.8520689 39.282630 42.674204 2 b_Aa2 14.6333534 1.1996304 12.239047 16.843446 3 b_Bb2 4.6176257 1.2040485 2.334325 7.039437 4 b_Bb3 -0.7612484 1.2100417 -3.033821 1.662261 5 b_Aa2:Bb2 -15.6839628 1.7100995 -18.928721 -12.285494 6 b_Aa2:Bb3 9.3101691 1.7191171 5.774495 12.433714 7 sigma 2.6429857 0.2638466 2.158565 3.141588
MCMC diagnostics
In addition to the regular model diagnostic checks (such as residual plots), for Bayesian analyses, it is necessary to explore the characteristics of the MCMC chains and the sampler in general. Recall that the purpose of MCMC sampling is to replicate the posterior distribution of the model likelihood and priors by drawing a known number of samples from this posterior (thereby formulating a probability distribution). This is only reliable if the MCMC samples accurately reflect the posterior.
Unfortunately, since we only know the posterior in the most trivial of circumstances, it is necessary to rely on indirect measures of how accurately the MCMC samples are likely to reflect the likelihood. I will breifly outline the most important diagnostics, however, please refer to Tutorial 4.3, Secton 3.1: Markov Chain Monte Carlo sampling for a discussion of these diagnostics.
- Traceplots for each parameter illustrate the MCMC sample values after each successive
iteration along the chain. Bad chain mixing (characterized by any sort of pattern) suggests
that the MCMC sampling chains may not have completely traversed all features of the posterior
distribution and that more iterations are required to ensure the distribution has been accurately
represented.

- Autocorrelation plot for each paramter illustrate the degree of correlation between
MCMC samples separated by different lags. For example, a lag of 0 represents the degree of
correlation between each MCMC sample and itself (obviously this will be a correlation of 1).
A lag of 1 represents the degree of correlation between each MCMC sample and the next sample along the Chain
and so on. In order to be able to generate unbiased estimates of parameters, the MCMC samples should be
independent (uncorrelated). In the figures below, this would be violated in the top autocorrelation plot and met in the bottom
autocorrelation plot.


- Rhat statistic for each parameter provides a measure of sampling efficiency/effectiveness. Ideally, all values should be less than 1.05. If there are values of 1.05 or greater it suggests that the sampler was not very efficient or effective. Not only does this mean that the sampler was potentiall slower than it could have been, more importantly, it could indicate that the sampler spent time sampling in a region of the likelihood that is less informative. Such a situation can arise from either a misspecified model or overly vague priors that permit sampling in otherwise nonscence parameter space.
Prior to inspecting any summaries of the parameter estimates, it is prudent to inspect a range of chain convergence diagnostics.
- Trace plots
View trace plotsTrace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
library(MCMCpack) plot(data.mcmcpack)


- Raftery diagnostic
View Raftery diagnosticThe Raftery diagnostics estimate that we would require about 3900 samples to reach the specified level of confidence in convergence. As we have 10,000 samples, we can be confidence that convergence has occurred.
library(MCMCpack) raftery.diag(data.mcmcpack)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) (Intercept) 2 3962 3746 1.060 Aa2 2 3620 3746 0.966 Bb2 2 3650 3746 0.974 Bb3 2 3771 3746 1.010 Aa2:Bb2 2 3865 3746 1.030 Aa2:Bb3 2 3741 3746 0.999 sigma2 2 3962 3746 1.060 - Autocorrelation diagnostic
View autocorrelationsA lag of 1 appears to be mainly sufficient to avoid autocorrelation.
library(MCMCpack) autocorr.diag(data.mcmcpack)
(Intercept) Aa2 Bb2 Bb3 Aa2:Bb2 Aa2:Bb3 sigma2 Lag 0 1.000000000 1.000000000 1.000000000 1.0000000000 1.000000000 1.000000000 1.000000000 Lag 1 -0.004894719 -0.012641557 -0.005109851 0.0087851394 -0.013378761 0.003611532 0.108486310 Lag 5 0.015615547 0.020010786 -0.008051140 0.0007380127 0.008677204 0.011141740 -0.003758313 Lag 10 0.012051793 0.023302011 -0.003096659 -0.0042219123 0.011092359 0.011419096 0.024786573 Lag 50 -0.002205204 -0.009273217 0.003096931 -0.0124255527 -0.019278730 -0.009166804 0.005734623
Again, prior to examining the summaries, we should have explored the convergence diagnostics.
library(coda) data.mcmc = as.mcmc(data.r2jags)
- Trace plots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
plot(data.mcmc)


When there are a lot of parameters, this can result in a very large number of traceplots. To focus on just certain parameters (such as $\beta$s)
data.mcmc = as.mcmc(data.r2jags) preds <- grep("beta", colnames(data.mcmc[[1]])) plot(data.mcmc[, preds])


- Raftery diagnostic
The Raftery diagnostics for each chain estimate that we would require no more than 5000 samples to reach the specified level of confidence in convergence. As we have 16,667 samples, we can be confidence that convergence has occurred.
raftery.diag(data.mcmc)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 38660 3746 10.30 beta[2] 20 37410 3746 9.99 beta[3] 20 38030 3746 10.20 beta[4] 20 36800 3746 9.82 beta[5] 20 36800 3746 9.82 beta[6] 20 35610 3746 9.51 deviance 20 36810 3746 9.83 sigma 20 38030 3746 10.20 [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 38660 3746 10.30 beta[2] 20 36800 3746 9.82 beta[3] 20 36800 3746 9.82 beta[4] 20 35610 3746 9.51 beta[5] 20 37410 3746 9.99 beta[6] 20 36800 3746 9.82 deviance 20 37410 3746 9.99 sigma 20 38030 3746 10.20 [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 38660 3746 10.30 beta[2] 20 38660 3746 10.30 beta[3] 20 37410 3746 9.99 beta[4] 20 39300 3746 10.50 beta[5] 20 36800 3746 9.82 beta[6] 20 36200 3746 9.66 deviance 20 37410 3746 9.99 sigma 20 37410 3746 9.99 - Autocorrelation diagnostic
A lag of 10 appears to be sufficient to avoid autocorrelation (poor mixing).
autocorr.diag(data.mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] deviance Lag 0 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 1.0000000000 1.0000000000 Lag 10 0.022642183 0.007087573 0.015327392 0.016544619 0.009691909 0.0127083887 0.0004530568 Lag 50 -0.005155744 0.006008383 0.001204183 -0.003596475 0.015488010 0.0008986928 0.0117358545 Lag 100 -0.010587005 0.014977295 -0.006923069 -0.008110816 -0.002500236 0.0059617664 0.0011406222 Lag 500 0.002228922 -0.001740023 -0.004237329 -0.002055156 0.006778323 -0.0046455847 0.0169645325 sigma Lag 0 1.0000000000 Lag 10 0.0111103826 Lag 50 -0.0003064979 Lag 100 0.0010915518 Lag 500 0.0063924921
Again, prior to examining the summaries, we should have explored the convergence diagnostics. There are numerous ways of working with STAN model fits (for exploring diagnostics and summarization).
- extract the mcmc samples and convert them into a mcmc.list to leverage the various coda routines
- use the numerous routines that come with the rstan package
- use the routines that come with the bayesplot package
- explore the diagnostics interactively via shinystan
- via coda
- Traceplots
- Autocorrelation
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.library(coda) s = as.array(data.rstan) wch = grep("beta", dimnames(s)$parameters) s = s[, , wch] mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)

 Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.library(coda) s = as.array(data.rstan) wch = grep("beta", dimnames(s)$parameters) s = s[, , wch] mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) autocorr.diag(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] Lag 0 1.00000000 1.000000000 1.00000000 1.000000000 1.00000000 Lag 1 0.14383604 0.176608647 0.12964311 0.126322833 0.13048872 Lag 5 -0.03522429 0.009474199 -0.05235367 -0.025111374 -0.01157408 Lag 10 0.01753957 -0.021221896 0.00910290 0.004300764 -0.02599045 Lag 50 0.02127630 0.036706859 0.01003985 -0.032431129 0.01426767
- via rstan
- Traceplots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
stan_trace(data.rstan)

- Raftery diagnostic
The Raftery diagnostics for each chain estimate that we would require no more than 5000 samples to reach the specified level of confidence in convergence. As we have 16,667 samples, we can be confidence that convergence has occurred.
raftery.diag(data.rstan)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Autocorrelation diagnostic
A lag of 2 appears broadly sufficient to avoid autocorrelation (poor mixing).
stan_ac(data.rstan)

- Rhat values. These values are a measure of sampling efficiency/effectiveness. Ideally, all values should be less than 1.05.
If there are values of 1.05 or greater it suggests that the sampler was not very efficient or effective. Not only does this
mean that the sampler was potentiall slower than it could have been, more importantly, it could indicate that the sampler spent time sampling
in a region of the likelihood that is less informative. Such a situation can arise from either a misspecified model or
overly vague priors that permit sampling in otherwise nonscence parameter space.
In this instance, all rhat values are well below 1.05 (a good thing).
stan_rhat(data.rstan)

- Another measure of sampling efficiency is Effective Sample Size (ess).
ess indicate the number samples (or proportion of samples that the sampling algorithm deamed effective. The sampler rejects samples
on the basis of certain criterion and when it does so, the previous sample value is used. Hence while the MCMC sampling chain
may contain 1000 samples, if there are only 10 effective samples (1%), the estimated properties are not likely to be reliable.
In this instance, most of the parameters have reasonably high effective samples and thus there is likely to be a good range of values from which to estimate paramter properties.
stan_ess(data.rstan)

- Traceplots
- via bayesplot
- Trace plots and density plots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
library(bayesplot) mcmc_trace(as.matrix(data.rstan), regex_pars = "beta|sigma")

library(bayesplot) mcmc_combo(as.matrix(data.rstan), regex_pars = "beta|sigma")

- Density plots
Density plots sugggest mean or median would be appropriate to describe the fixed posteriors and median is appropriate for the sigma posterior.
library(bayesplot) mcmc_dens(as.matrix(data.rstan), regex_pars = "beta|sigma")

- Trace plots and density plots
- via shinystan
library(shinystan) launch_shinystan(data.rstan)
- It is worth exploring the influence of our priors.
Again, prior to examining the summaries, we should have explored the convergence diagnostics. There are numerous ways of working with STANARM model fits (for exploring diagnostics and summarization).
- extract the mcmc samples and convert them into a mcmc.list to leverage the various coda routines
- use the numerous routines that come with the rstan package
- use the routines that come with the bayesplot package
- explore the diagnostics interactively via shinystan
- via coda
- Traceplots
- Autocorrelation
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.library(coda) s = as.array(data.rstanarm) mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)

 Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.library(coda) s = as.array(data.rstanarm) mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) autocorr.diag(mcmc)
(Intercept) Aa2 Bb2 Bb3 Aa2:Bb2 Aa2:Bb3 Lag 0 1.000000000 1.0000000000 1.00000000 1.0000000000 1.000000000 1.000000000 Lag 1 0.079250889 0.0525242478 0.02921679 0.0118875596 0.019013378 0.050538090 Lag 5 0.016286755 0.0210690165 -0.01755251 -0.0006499424 -0.001479584 0.019445419 Lag 10 -0.003125278 -0.0058528012 -0.01445239 -0.0308228680 -0.022582741 -0.009611371 Lag 50 -0.001455135 -0.0002293142 0.03901632 -0.0366835504 0.048671623 -0.001621902
- via rstan
- Traceplots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
stan_trace(data.rstanarm)

- Raftery diagnostic
The Raftery diagnostics for each chain estimate that we would require no more than 5000 samples to reach the specified level of confidence in convergence. As we have 16,667 samples, we can be confidence that convergence has occurred.
raftery.diag(data.rstanarm)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Autocorrelation diagnostic
A lag of 2 appears broadly sufficient to avoid autocorrelation (poor mixing).
stan_ac(data.rstanarm)

- Rhat values. These values are a measure of sampling efficiency/effectiveness. Ideally, all values should be less than 1.05.
If there are values of 1.05 or greater it suggests that the sampler was not very efficient or effective. Not only does this
mean that the sampler was potentiall slower than it could have been, more importantly, it could indicate that the sampler spent time sampling
in a region of the likelihood that is less informative. Such a situation can arise from either a misspecified model or
overly vague priors that permit sampling in otherwise nonscence parameter space.
In this instance, all rhat values are well below 1.05 (a good thing).
stan_rhat(data.rstanarm)

- Another measure of sampling efficiency is Effective Sample Size (ess).
ess indicate the number samples (or proportion of samples that the sampling algorithm deamed effective. The sampler rejects samples
on the basis of certain criterion and when it does so, the previous sample value is used. Hence while the MCMC sampling chain
may contain 1000 samples, if there are only 10 effective samples (1%), the estimated properties are not likely to be reliable.
In this instance, most of the parameters have reasonably high effective samples and thus there is likely to be a good range of values from which to estimate paramter properties.
stan_ess(data.rstanarm)

- Traceplots
- via bayesplot
- Trace plots and density plots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
library(bayesplot) mcmc_trace(as.array(data.rstanarm), regex_pars = "Intercept|x|sigma")

mcmc_combo(as.array(data.rstanarm))

- Density plots
Density plots sugggest mean or median would be appropriate to describe the fixed posteriors and median is appropriate for the sigma posterior.
mcmc_dens(as.array(data.rstanarm))

- Trace plots and density plots
- via rstanarm
The rstanarm package provides additional posterior checks.- Posterior vs Prior - this compares the posterior estimate for each parameter against the associated prior.
If the spread of the priors is small relative to the posterior, then it is likely that the priors are too influential.
On the other hand, overly wide priors can lead to computational issues.
library(rstanarm) posterior_vs_prior(data.rstanarm, color_by = "vs", group_by = TRUE, facet_args = list(scales = "free_y"))
Gradient evaluation took 3.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.35 seconds. Adjust your expectations accordingly! Elapsed Time: 0.287642 seconds (Warm-up) 0.12345 seconds (Sampling) 0.411092 seconds (Total) Gradient evaluation took 1.3e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.13 seconds. Adjust your expectations accordingly! Elapsed Time: 0.262367 seconds (Warm-up) 0.1157 seconds (Sampling) 0.378067 seconds (Total)
- Posterior vs Prior - this compares the posterior estimate for each parameter against the associated prior.
If the spread of the priors is small relative to the posterior, then it is likely that the priors are too influential.
On the other hand, overly wide priors can lead to computational issues.
- via shinystan
library(shinystan) launch_shinystan(data.rstanarm)
Again, prior to examining the summaries, we should have explored the convergence diagnostics. Rather than dublicate this for both additive and multiplicative models, we will only explore the multiplicative model. There are numerous ways of working with STAN model fits (for exploring diagnostics and summarization).
- extract the mcmc samples and convert them into a mcmc.list to leverage the various coda routines
- use the numerous routines that come with the rstan package
- use the routines that come with the bayesplot package
- explore the diagnostics interactively via shinystan
- via coda
- Traceplots
- Autocorrelation
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.library(coda) mcmc = as.mcmc(data.brms) plot(mcmc)

 Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.library(coda) mcmc = as.mcmc(data.brms) autocorr.diag(mcmc)
Error in ts(x, start = start(x), end = end(x), deltat = thin(x)): invalid time series parameters specified
- via rstan
- Traceplots
Trace plots show no evidence that the chains have not reasonably traversed the entire multidimensional parameter space.
stan_trace(data.brms$fit)

- Raftery diagnostic
The Raftery diagnostics for each chain estimate that we would require no more than 5000 samples to reach the specified level of confidence in convergence. As we have 16,667 samples, we can be confidence that convergence has occurred.
raftery.diag(data.brms)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
- Autocorrelation diagnostic
A lag of 2 appears broadly sufficient to avoid autocorrelation (poor mixing).
stan_ac(data.brms$fit)

- Rhat values. These values are a measure of sampling efficiency/effectiveness. Ideally, all values should be less than 1.05.
If there are values of 1.05 or greater it suggests that the sampler was not very efficient or effective. Not only does this
mean that the sampler was potentiall slower than it could have been, more importantly, it could indicate that the sampler spent time sampling
in a region of the likelihood that is less informative. Such a situation can arise from either a misspecified model or
overly vague priors that permit sampling in otherwise nonscence parameter space.
In this instance, all rhat values are well below 1.05 (a good thing).
stan_rhat(data.brms$fit)

- Another measure of sampling efficiency is Effective Sample Size (ess).
ess indicate the number samples (or proportion of samples that the sampling algorithm deamed effective. The sampler rejects samples
on the basis of certain criterion and when it does so, the previous sample value is used. Hence while the MCMC sampling chain
may contain 1000 samples, if there are only 10 effective samples (1%), the estimated properties are not likely to be reliable.
In this instance, most of the parameters have reasonably high effective samples and thus there is likely to be a good range of values from which to estimate paramter properties.
stan_ess(data.brms$fit)

- Traceplots
Model validation
Model validation involves exploring the model diagnostics and fit to ensure that the model is broadly appropriate for the data. As such, exploration of the residuals should be routine.
For more complex models (those that contain multiple effects, it is also advisable to plot the residuals against each of the individual predictors. For sampling designs that involve sample collection over space or time, it is also a good idea to explore whether there are any temporal or spatial patterns in the residuals.
There are numerous situations (e.g. when applying specific variance-covariance structures to a model) where raw residuals do not reflect the interior workings of the model. Typically, this is because they do not take into account the variance-covariance matrix or assume a very simple variance-covariance matrix. Since the purpose of exploring residuals is to evaluate the model, for these cases, it is arguably better to draw conclusions based on standardized (or studentized) residuals.
Unfortunately the definitions of standardized and studentized residuals appears to vary and the two terms get used interchangeably. I will adopt the following definitions:
| Standardized residuals: | the raw residuals divided by the true standard deviation of the residuals (which of course is rarely known). | |
| Studentized residuals: | the raw residuals divided by the standard deviation of the residuals. Note that externally studentized residuals are calculated by dividing the raw residuals by a unique standard deviation for each observation that is calculated from regressions having left each successive observation out. | |
| Pearson residuals: | the raw residuals divided by the standard deviation of the response variable. |
The mark of a good model is being able to predict well. In an ideal world, we would have sufficiently large sample size as to permit us to hold a fraction (such as 25%) back thereby allowing us to train the model on 75% of the data and then see how well the model can predict the withheld 25%. Unfortunately, such a luxury is still rare in ecology.
The next best option is to see how well the model can predict the observed data. Models tend to struggle most with the extremes of trends and have particular issues when the extremes approach logical boundaries (such as zero for count data and standard deviations). We can use the fitted model to generate random predicted observations and then explore some properties of these compared to the actual observed data.
Residuals are not computed directly within MCMCpack. However, we can calculate them manually form the posteriors.
mcmc = as.data.frame(data.mcmcpack) # generate a model matrix newdata = data Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates head(mcmc)
(Intercept) Aa2 Bb2 Bb3 Aa2:Bb2 Aa2:Bb3 sigma2 1 40.42172 14.17136 3.941759 -0.3299995 -14.06168 10.470534 5.338679 2 41.37944 14.82348 4.396285 -2.2414088 -16.73676 9.835985 8.130920 3 40.90443 14.60643 3.996873 -2.1078572 -12.63888 9.953201 7.456621 4 41.13543 15.74346 5.006105 -1.1591951 -18.15799 9.737565 8.063221 5 41.09695 15.74244 4.816085 0.2531722 -16.46468 7.425350 6.836169 6 40.77523 15.04709 5.929086 0.7056246 -16.61962 8.402492 5.451654
wch = grepl("sigma2", colnames(mcmc)) == 0 coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

Residuals against predictors
mcmc = as.data.frame(data.mcmcpack) # generate a model matrix newdata = newdata Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates wch = grepl("sigma", colnames(mcmc)) == 0 coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit newdata = newdata %>% cbind(fit, resid) ggplot(newdata) + geom_point(aes(y = resid, x = A))

ggplot(newdata) + geom_point(aes(y = resid, x = B))

And now for studentized residuals
mcmc = as.data.frame(data.mcmcpack) # generate a model matrix newdata = data Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates wch = grepl("sigma", colnames(mcmc)) == 0 coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit sresid = resid/sd(resid) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

Conclusions: for this simple model, the studentized residuals yield the same pattern as the raw residuals (or the Pearson residuals for that matter).
Lets see how well data simulated from the model reflects the raw data
mcmc = as.matrix(data.mcmcpack) #generate a model matrix Xmat = model.matrix(~A*B, data) ##get median parameter estimates wch = grepl('sigma',colnames(mcmc))==0 coefs = mcmc[,wch] fit = coefs %*% t(Xmat) ## draw samples from this model yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(data), fit[i,], sqrt(mcmc[i, 'sigma2']))) newdata = data.frame(A=data$A, B=data$B, yRep) %>% gather(key=Sample, value=Value,-A,-B) ggplot(newdata) + geom_violin(aes(y=Value, x=A, fill='Model'), alpha=0.5)+ geom_violin(data=data, aes(y=y,x=A,fill='Obs'), alpha=0.5) + geom_point(data=data, aes(y=y, x=A), position=position_jitter(width=0.1,height=0), color='black')

ggplot(newdata) + geom_violin(aes(y=Value, x=B, fill='Model', group=B, color=A), alpha=0.5)+ geom_point(data=data, aes(y=y, x=B, group=B,color=A))

Conclusions the predicted trends do encapsulate the actual data, suggesting that the model is a reasonable representation of the underlying processes. Note, these are prediction intervals rather than confidence intervals as we are seeking intervals within which we can predict individual observations rather than means.
We can also explore the posteriors of each parameter.
library(bayesplot) mcmc_intervals(as.matrix(data.mcmcpack), regex_pars = "Intercept|^A|^B|sigma")

mcmc_areas(as.matrix(data.mcmcpack), regex_pars = "Intercept|^A|^B|sigma")

Residuals are not computed directly within JAGS. However, we can calculate them manually form the posteriors.
mcmc = data.r2jags$BUGSoutput$sims.matrix # generate a model matrix newdata = data Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates wch = grep("beta\\[", colnames(mcmc)) wch
[1] 1 2 3 4 5 6
head(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] deviance sigma [1,] 40.48339 15.06364 5.935638 -0.3093790 -17.87132 8.043827 283.5346 2.538900 [2,] 41.63781 14.36993 1.931867 -1.1518205 -13.69143 9.739930 286.9971 2.768452 [3,] 40.61291 14.92807 4.636804 -0.4964191 -15.13200 10.374972 289.3339 3.337160 [4,] 42.04693 14.50413 5.536484 -2.2035782 -17.60762 8.681790 289.7733 2.488243 [5,] 42.38741 12.94789 4.119175 -0.8898141 -14.22881 11.716873 289.7752 2.617655 [6,] 40.14774 15.40453 4.608544 2.2212448 -16.07778 6.200402 288.8282 2.851959
coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

Residuals against predictors
mcmc = data.r2jags$BUGSoutput$sims.matrix wch = grep("beta\\[", colnames(mcmc)) # generate a model matrix newdata = newdata Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates coefs = apply(mcmc[, wch], 2, median) print(coefs)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] 41.0945676 14.6518796 4.6535912 -0.7356942 -15.7214406 9.3350177
fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit newdata = newdata %>% cbind(fit, resid) ggplot(newdata) + geom_point(aes(y = resid, x = A))

ggplot(newdata) + geom_point(aes(y = resid, x = B))

And now for studentized residuals
mcmc = data.r2jags$BUGSoutput$sims.matrix wch = grep("beta\\[", colnames(mcmc)) # generate a model matrix newdata = data Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit sresid = resid/sd(resid) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

Conclusions: for this simple model, the studentized residuals yield the same pattern as the raw residuals (or the Pearson residuals for that matter).
Lets see how well data simulated from the model reflects the raw data
mcmc = data.r2jags$BUGSoutput$sims.matrix wch = grep("beta\\[", colnames(mcmc)) #generate a model matrix Xmat = model.matrix(~A*B, data) ##get median parameter estimates coefs = mcmc[,wch] fit = coefs %*% t(Xmat) ## draw samples from this model yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(data), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(A=data$A, B=data$B, yRep) %>% gather(key=Sample, value=Value,-A,-B) ggplot(newdata) + geom_violin(aes(y=Value, x=A, fill='Model'), alpha=0.5)+ geom_violin(data=data, aes(y=y,x=A,fill='Obs'), alpha=0.5) + geom_point(data=data, aes(y=y, x=A), position=position_jitter(width=0.1,height=0), color='black')

ggplot(newdata) + geom_violin(aes(y=Value, x=B, fill='Model', group=B, color=A), alpha=0.5)+ geom_point(data=data, aes(y=y, x=B, group=B,color=A))

Conclusions the predicted trends do encapsulate the actual data, suggesting that the model is a reasonable representation of the underlying processes. Note, these are prediction intervals rather than confidence intervals as we are seeking intervals within which we can predict individual observations rather than means.
We can also explore the posteriors of each parameter.
library(bayesplot) mcmc_intervals(data.r2jags$BUGSoutput$sims.matrix, regex_pars = "beta|sigma")

mcmc_areas(data.r2jags$BUGSoutput$sims.matrix, regex_pars = "beta|sigma")

Residuals are not computed directly within STAN. However, we can calculate them manually form the posteriors.
mcmc = as.matrix(data.rstan) # generate a model matrix newdata = data Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates wch = grep("beta\\[", colnames(mcmc)) coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

Residuals against predictors
mcmc = as.matrix(data.rstan) wch = grep("beta\\[", colnames(mcmc)) # generate a model matrix newdata = newdata Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates coefs = apply(mcmc[, wch], 2, median) print(coefs)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] 41.1578967 14.6211877 4.5876538 -0.8094151 -15.6448600 9.3816379
fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit newdata = newdata %>% cbind(fit, resid) ggplot(newdata) + geom_point(aes(y = resid, x = A))

ggplot(newdata) + geom_point(aes(y = resid, x = B))

And now for studentized residuals
mcmc = as.matrix(data.rstan) wch = grep("beta\\[", colnames(mcmc)) # generate a model matrix newdata = data Xmat = model.matrix(~A * B, newdata) ## get median parameter estimates coefs = apply(mcmc[, wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = data$y - fit sresid = resid/sd(resid) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

Conclusions: for this simple model, the studentized residuals yield the same pattern as the raw residuals (or the Pearson residuals for that matter).
Lets see how well data simulated from the model reflects the raw data
mcmc = as.matrix(data.rstan) wch = grep("beta\\[", colnames(mcmc)) #generate a model matrix Xmat = model.matrix(~A*B, data) ##get median parameter estimates coefs = mcmc[,wch] fit = coefs %*% t(Xmat) ## draw samples from this model yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(data), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(A=data$A, B=data$B, yRep) %>% gather(key=Sample, value=Value,-A,-B) ggplot(newdata) + geom_violin(aes(y=Value, x=A, fill='Model'), alpha=0.5)+ geom_violin(data=data, aes(y=y,x=A,fill='Obs'), alpha=0.5) + geom_point(data=data, aes(y=y, x=A), position=position_jitter(width=0.1,height=0), color='black')

ggplot(newdata) + geom_violin(aes(y=Value, x=B, fill='Model', group=B, color=A), alpha=0.5)+ geom_point(data=data, aes(y=y, x=B, group=B,color=A))

Conclusions the predicted trends do encapsulate the actual data, suggesting that the model is a reasonable representation of the underlying processes. Note, these are prediction intervals rather than confidence intervals as we are seeking intervals within which we can predict individual observations rather than means.
We can also explore the posteriors of each parameter.
library(bayesplot) mcmc_intervals(as.matrix(data.rstan), regex_pars = "beta|sigma")

mcmc_areas(as.matrix(data.rstan), regex_pars = "beta|sigma")

Residuals are not computed directly within RSTANARM. However, we can calculate them manually form the posteriors.
resid = resid(data.rstanarm) fit = fitted(data.rstanarm) ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

Residuals against predictors
resid = resid(data.rstanarm) dat = data %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = A))

ggplot(dat) + geom_point(aes(y = resid, x = B))

And now for studentized residuals
resid = resid(data.rstanarm) sigma(data.rstanarm)
[1] 2.626127
sresid = resid/sigma(data.rstanarm) fit = fitted(data.rstanarm) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

Conclusions: for this simple model, the studentized residuals yield the same pattern as the raw residuals (or the Pearson residuals for that matter).
Lets see how well data simulated from the model reflects the raw data
y_pred = posterior_predict(data.rstanarm) newdata = data %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -A,-B,-y) ggplot(newdata) + geom_violin(aes(y = Value, x = A, fill = "Model"),alpha = 0.5) + geom_violin(data = data, aes(y = y, x = A,fill = "Obs"), alpha = 0.5) + geom_point(data = data, aes(y = y,x = A), position = position_jitter(width = 0.1, height = 0), color = "black")

ggplot(newdata) + geom_violin(aes(y=Value, x=B, fill='Model', group=B, color=A), alpha=0.5)+ geom_point(data=data, aes(y=y, x=B, group=B,color=A))

Conclusions the predicted trends do encapsulate the actual data, suggesting that the model is a reasonable representation of the underlying processes. Note, these are prediction intervals rather than confidence intervals as we are seeking intervals within which we can predict individual observations rather than means.
We can also explore the posteriors of each parameter.
library(bayesplot) mcmc_intervals(as.matrix(data.rstanarm), regex_pars = "Intercept|^A|^B|sigma")

mcmc_areas(as.matrix(data.rstanarm), regex_pars = "Intercept|^A|^B|sigma")

Residuals are not computed directly within BRMS. However, we can calculate them manually form the posteriors.
resid = resid(data.brms)[, "Estimate"] fit = fitted(data.brms)[, "Estimate"] ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

Residuals against predictors
resid = resid(data.brms)[, "Estimate"] dat = data %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = A))

ggplot(dat) + geom_point(aes(y = resid, x = B))

And now for studentized residuals
resid = resid(data.brms)[, "Estimate"] sresid = resid(data.brms, type = "pearson")[, "Estimate"] fit = fitted(data.brms)[, "Estimate"] ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

Conclusions: for this simple model, the studentized residuals yield the same pattern as the raw residuals (or the Pearson residuals for that matter).
Lets see how well data simulated from the model reflects the raw data
y_pred = posterior_predict(data.brms) newdata = data %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -A,-B,-y) ggplot(newdata) + geom_violin(aes(y = Value, x = A, fill = "Model"),alpha = 0.5) + geom_violin(data = data, aes(y = y, x = A,fill = "Obs"), alpha = 0.5) + geom_point(data = data, aes(y = y,x = A), position = position_jitter(width = 0.1, height = 0), color = "black")

ggplot(newdata) + geom_violin(aes(y=Value, x=B, fill='Model', group=B, color=A), alpha=0.5)+ geom_point(data=data, aes(y=y, x=B, group=B,color=A))

Conclusions the predicted trends do encapsulate the actual data, suggesting that the model is a reasonable representation of the underlying processes. Note, these are prediction intervals rather than confidence intervals as we are seeking intervals within which we can predict individual observations rather than means.
We can also explore the posteriors of each parameter.
library(bayesplot) mcmc_intervals(as.matrix(data.brms), regex_pars = "Intercept|b_|sigma")

mcmc_areas(as.matrix(data.brms), regex_pars = "Intercept|b_|sigma")

Parameter estimates (posterior summaries)
Although all parameters in a Bayesian analysis are considered random and are considered a distribution, rarely would it be useful to present tables of all the samples from each distribution. On the other hand, plots of the posterior distributions are do have some use. Nevertheless, most workers prefer to present simple statistical summaries of the posteriors. Popular choices include the median (or mean) and 95% credibility intervals.
library(coda) mcmcpvalue <- function(samp) { ## elementary version that creates an empirical p-value for the ## hypothesis that the columns of samp have mean zero versus a general ## multivariate distribution with elliptical contours. ## differences from the mean standardized by the observed ## variance-covariance factor ## Note, I put in the bit for single terms if (length(dim(samp)) == 0) { std <- backsolve(chol(var(samp)), cbind(0, t(samp)) - mean(samp), transpose = TRUE) sqdist <- colSums(std * std) sum(sqdist[-1] > sqdist[1])/length(samp) } else { std <- backsolve(chol(var(samp)), cbind(0, t(samp)) - colMeans(samp), transpose = TRUE) sqdist <- colSums(std * std) sum(sqdist[-1] > sqdist[1])/nrow(samp) } }
Matrix model (MCMCpack)
summary(data.mcmcpack)
Iterations = 1001:11000
Thinning interval = 1
Number of chains = 1
Sample size per chain = 10000
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
(Intercept) 41.103 0.8453 0.008453 0.008513
Aa2 14.645 1.1977 0.011977 0.011977
Bb2 4.638 1.1843 0.011843 0.011843
Bb3 -0.767 1.1920 0.011920 0.011920
Aa2:Bb2 -15.714 1.6826 0.016826 0.016923
Aa2:Bb3 9.350 1.6806 0.016806 0.016806
sigma2 7.019 1.4188 0.014188 0.015822
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
(Intercept) 39.456 40.534 41.1032 41.66037 42.778
Aa2 12.270 13.844 14.6562 15.44678 16.996
Bb2 2.325 3.844 4.6501 5.42905 6.966
Bb3 -3.122 -1.547 -0.7767 0.03978 1.564
Aa2:Bb2 -19.008 -16.846 -15.7026 -14.57069 -12.477
Aa2:Bb3 6.017 8.231 9.3582 10.47091 12.658
sigma2 4.759 6.006 6.8646 7.83681 10.312
# OR library(broom) tidyMCMC(data.mcmcpack, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 (Intercept) 41.1032161 0.8453382 39.473479 42.784450 2 Aa2 14.6449428 1.1976886 12.238706 16.934341 3 Bb2 4.6379857 1.1843355 2.219722 6.842731 4 Bb3 -0.7670406 1.1920203 -3.141803 1.531671 5 Aa2:Bb2 -15.7143139 1.6825773 -18.991196 -12.467581 6 Aa2:Bb3 9.3499949 1.6805981 6.073684 12.710434 7 sigma2 7.0191843 1.4188454 4.563271 9.892696
- the intercept represents the mean of the first combination Aa1:Bb1 is
41.1032161 - Aa2:Bb1 is
14.6449428units greater than Aa1:Bb1 - Aa1:Bb2 is
4.6379857units greater Aa1:Bb1 - Aa1:Bb3 is
-0.7670406units greater Aa1:Bb1 - Aa2:Bb2 is
-15.7143139units greater than the difference between (Aa1:Bb2 + Aa2:Bb1) and (2*Aa1:Bb1) - Aa2:Bb3 is
9.3499949units greater than the difference between (Aa1:Bb3 + Aa2:Bb1) and (2*Aa1:Bb1)
While workers attempt to become comfortable with a new statistical framework, it is only natural that they like to evaluate and comprehend new structures and output alongside more familiar concepts. One way to facilitate this is via Bayesian p-values that are somewhat analogous to the frequentist p-values for investigating the hypothesis that a parameter is equal to zero.
## since values are less than zero mcmcpvalue(data.mcmcpack[, 2])
[1] 0
mcmcpvalue(data.mcmcpack[, 3])
[1] 4e-04
mcmcpvalue(data.mcmcpack[, 4])
[1] 0.5129
mcmcpvalue(data.mcmcpack[, 5])
[1] 0
mcmcpvalue(data.mcmcpack[, 6])
[1] 0
mcmcpvalue(data.mcmcpack[, 5:6])
[1] 0
There is evidence of an interaction between A and B.
Matrix model (JAGS)
print(data.r2jags)
Inference for Bugs model at "5", fit using jags,
3 chains, each with 53000 iterations (first 3000 discarded), n.thin = 10
n.sims = 15000 iterations saved
mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff
beta[1] 41.094 0.844 39.441 40.530 41.095 41.652 42.743 1.001 15000
beta[2] 14.647 1.194 12.274 13.867 14.652 15.442 16.982 1.001 15000
beta[3] 4.652 1.193 2.295 3.852 4.654 5.441 6.977 1.001 15000
beta[4] -0.746 1.190 -3.126 -1.515 -0.736 0.046 1.597 1.001 15000
beta[5] -15.712 1.695 -19.045 -16.834 -15.721 -14.601 -12.325 1.001 15000
beta[6] 9.336 1.673 6.039 8.232 9.335 10.434 12.585 1.001 15000
sigma 2.662 0.261 2.210 2.478 2.639 2.826 3.227 1.001 15000
deviance 286.111 4.072 280.390 283.115 285.406 288.308 295.927 1.001 15000
For each parameter, n.eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor (at convergence, Rhat=1).
DIC info (using the rule, pD = var(deviance)/2)
pD = 8.3 and DIC = 294.4
DIC is an estimate of expected predictive error (lower deviance is better).
# OR library(broom) tidyMCMC(as.mcmc(data.r2jags), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 beta[1] 41.0939179 0.8444629 39.424828 42.720949 2 beta[2] 14.6468728 1.1936367 12.362757 17.040371 3 beta[3] 4.6516490 1.1925610 2.296748 6.978871 4 beta[4] -0.7456183 1.1896540 -3.175750 1.543696 5 beta[5] -15.7117566 1.6948094 -19.140973 -12.442649 6 beta[6] 9.3364992 1.6732387 6.034525 12.572229 7 deviance 286.1113628 4.0724442 279.611182 294.244142 8 sigma 2.6621371 0.2610362 2.193585 3.199500
- the intercept represents the mean of the first combination Aa1:Bb1 is
41.0939179 - Aa2:Bb1 is
14.6468728units greater than Aa1:Bb1 - Aa1:Bb2 is
4.651649units greater Aa1:Bb1 - Aa1:Bb3 is
-0.7456183units greater Aa1:Bb1 - Aa2:Bb2 is
-15.7117566units greater than the difference between (Aa1:Bb2 + Aa2:Bb1) and (2*Aa1:Bb1) - Aa2:Bb3 is
9.3364992units greater than the difference between (Aa1:Bb3 + Aa2:Bb1) and (2*Aa1:Bb1)
While workers attempt to become comfortable with a new statistical framework, it is only natural that they like to evaluate and comprehend new structures and output alongside more familiar concepts. One way to facilitate this is via Bayesian p-values that are somewhat analogous to the frequentist p-values for investigating the hypothesis that a parameter is equal to zero.
## since values are less than zero mcmcpvalue(data.r2jags$BUGSoutput$sims.matrix[, "beta[2]"])
[1] 0
mcmcpvalue(data.r2jags$BUGSoutput$sims.matrix[, "beta[3]"])
[1] 6.666667e-05
mcmcpvalue(data.r2jags$BUGSoutput$sims.matrix[, "beta[4]"])
[1] 0.5181333
mcmcpvalue(data.r2jags$BUGSoutput$sims.matrix[, "beta[5]"])
[1] 0
mcmcpvalue(data.r2jags$BUGSoutput$sims.matrix[, "beta[6]"])
[1] 0
mcmcpvalue(data.r2jags$BUGSoutput$sims.matrix[, c("beta[5]", "beta[6]")])
[1] 0
There is evidence of an interaction between A and B.
Matrix model (STAN)
print(data.rstan, pars = c("beta", "sigma"))
Inference for Stan model: 3d2414c9dcf4b5e12be870eadd2c894a.
3 chains, each with iter=2000; warmup=500; thin=3;
post-warmup draws per chain=500, total post-warmup draws=1500.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
beta[1] 41.13 0.03 0.84 39.47 40.54 41.16 41.68 42.74 1070 1
beta[2] 14.60 0.04 1.17 12.36 13.80 14.62 15.36 16.94 1026 1
beta[3] 4.60 0.04 1.20 2.28 3.81 4.59 5.40 6.89 1134 1
beta[4] -0.79 0.04 1.17 -3.07 -1.55 -0.81 -0.03 1.65 1065 1
beta[5] -15.64 0.05 1.61 -18.74 -16.74 -15.64 -14.51 -12.48 1141 1
beta[6] 9.38 0.05 1.63 6.22 8.27 9.38 10.51 12.69 1080 1
sigma 2.66 0.01 0.26 2.21 2.47 2.63 2.83 3.24 1232 1
Samples were drawn using NUTS(diag_e) at Sat Nov 25 17:19:17 2017.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
# OR library(broom) tidyMCMC(data.rstan, conf.int = TRUE, conf.method = "HPDinterval", pars = c("beta", "sigma"))
term estimate std.error conf.low conf.high 1 beta[1] 41.128049 0.8374676 39.596978 42.796440 2 beta[2] 14.604800 1.1720223 12.294697 16.856289 3 beta[3] 4.601844 1.1980414 2.314024 6.918298 4 beta[4] -0.790552 1.1684885 -3.102146 1.620022 5 beta[5] -15.639649 1.6141412 -18.956622 -12.847933 6 beta[6] 9.384906 1.6348109 6.033244 12.348575 7 sigma 2.661834 0.2635065 2.193934 3.195871
- the intercept represents the mean of the first combination Aa1:Bb1 is
41.1280486 - Aa2:Bb1 is
14.6047995units greater than Aa1:Bb1 - Aa1:Bb2 is
4.6018442units greater Aa1:Bb1 - Aa1:Bb3 is
-0.790552units greater Aa1:Bb1 - Aa2:Bb2 is
-15.6396486units greater than the difference between (Aa1:Bb2 + Aa2:Bb1) and (2*Aa1:Bb1) - Aa2:Bb3 is
9.3849055units greater than the difference between (Aa1:Bb3 + Aa2:Bb1) and (2*Aa1:Bb1)
While workers attempt to become comfortable with a new statistical framework, it is only natural that they like to evaluate and comprehend new structures and output alongside more familiar concepts. One way to facilitate this is via Bayesian p-values that are somewhat analogous to the frequentist p-values for investigating the hypothesis that a parameter is equal to zero.
## since values are less than zero mcmcpvalue(as.matrix(data.rstan)[, "beta[2]"])
[1] 0
mcmcpvalue(as.matrix(data.rstan)[, "beta[3]"])
[1] 0.0006666667
mcmcpvalue(as.matrix(data.rstan)[, "beta[4]"])
[1] 0.486
mcmcpvalue(as.matrix(data.rstan)[, "beta[5]"])
[1] 0
mcmcpvalue(as.matrix(data.rstan)[, "beta[6]"])
[1] 0
mcmcpvalue(as.matrix(data.rstan)[, c("beta[5]", "beta[6]")])
[1] 0
There is evidence of an interaction between A and B.
library(loo) (full = loo(extract_log_lik(data.rstan)))
Computed from 1500 by 60 log-likelihood matrix
Estimate SE
elpd_loo -146.8 6.1
p_loo 6.7 1.5
looic 293.7 12.2
Pareto k diagnostic values:
Count Pct
(-Inf, 0.5] (good) 59 98.3%
(0.5, 0.7] (ok) 1 1.7%
(0.7, 1] (bad) 0 0.0%
(1, Inf) (very bad) 0 0.0%
All Pareto k estimates are ok (k < 0.7)
See help('pareto-k-diagnostic') for details.
# now fit a model without main factor modelString = " data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } transformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,1000); sigma~cauchy(0,5); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " Xmat <- model.matrix(~A + B, data) data.list <- with(data, list(y = y, X = Xmat, n = nrow(data), nX = ncol(Xmat))) data.rstan.red <- stan(data = data.list, model_code = modelString, chains = 3, iter = 2000, warmup = 500, thin = 3)
In file included from /usr/local/lib/R/site-library/BH/include/boost/config.hpp:39:0,
from /usr/local/lib/R/site-library/BH/include/boost/math/tools/config.hpp:13,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/var.hpp:7,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/gevv_vvv_vari.hpp:5,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core.hpp:12,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/mat.hpp:4,
from /usr/local/lib/R/site-library/StanHeaders/include/stan/math.hpp:4,
from /usr/local/lib/R/site-library/StanHeaders/include/src/stan/model/model_header.hpp:4,
from file5e932ec268fb.cpp:8:
/usr/local/lib/R/site-library/BH/include/boost/config/compiler/gcc.hpp:186:0: warning: "BOOST_NO_CXX11_RVALUE_REFERENCES" redefined
# define BOOST_NO_CXX11_RVALUE_REFERENCES
^
<command-line>:0:0: note: this is the location of the previous definition
SAMPLING FOR MODEL '3b057d3d81cbed2078ce678376a94574' NOW (CHAIN 1).
Gradient evaluation took 6.2e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.62 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 501 / 2000 [ 25%] (Sampling)
Iteration: 700 / 2000 [ 35%] (Sampling)
Iteration: 900 / 2000 [ 45%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
Iteration: 1300 / 2000 [ 65%] (Sampling)
Iteration: 1500 / 2000 [ 75%] (Sampling)
Iteration: 1700 / 2000 [ 85%] (Sampling)
Iteration: 1900 / 2000 [ 95%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.110346 seconds (Warm-up)
0.18655 seconds (Sampling)
0.296896 seconds (Total)
SAMPLING FOR MODEL '3b057d3d81cbed2078ce678376a94574' NOW (CHAIN 2).
Gradient evaluation took 3.8e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.38 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 501 / 2000 [ 25%] (Sampling)
Iteration: 700 / 2000 [ 35%] (Sampling)
Iteration: 900 / 2000 [ 45%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
Iteration: 1300 / 2000 [ 65%] (Sampling)
Iteration: 1500 / 2000 [ 75%] (Sampling)
Iteration: 1700 / 2000 [ 85%] (Sampling)
Iteration: 1900 / 2000 [ 95%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.098975 seconds (Warm-up)
0.189632 seconds (Sampling)
0.288607 seconds (Total)
SAMPLING FOR MODEL '3b057d3d81cbed2078ce678376a94574' NOW (CHAIN 3).
Gradient evaluation took 4.8e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.48 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 501 / 2000 [ 25%] (Sampling)
Iteration: 700 / 2000 [ 35%] (Sampling)
Iteration: 900 / 2000 [ 45%] (Sampling)
Iteration: 1100 / 2000 [ 55%] (Sampling)
Iteration: 1300 / 2000 [ 65%] (Sampling)
Iteration: 1500 / 2000 [ 75%] (Sampling)
Iteration: 1700 / 2000 [ 85%] (Sampling)
Iteration: 1900 / 2000 [ 95%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.106341 seconds (Warm-up)
0.148901 seconds (Sampling)
0.255242 seconds (Total)
(reduced = loo(extract_log_lik(data.rstan.red)))
Computed from 1500 by 60 log-likelihood matrix
Estimate SE
elpd_loo -194.6 3.7
p_loo 4.2 0.5
looic 389.2 7.4
All Pareto k estimates are good (k < 0.5)
See help('pareto-k-diagnostic') for details.
par(mfrow = 1:2, mar = c(5, 3.8, 1, 0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full, reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
Matrix model (RSTANARM)
summary(data.rstanarm)
Model Info:
function: stan_glm
family: gaussian [identity]
formula: y ~ A * B
algorithm: sampling
priors: see help('prior_summary')
sample: 2250 (posterior sample size)
num obs: 60
Estimates:
mean sd 2.5% 25% 50% 75% 97.5%
(Intercept) 41.1 0.8 39.4 40.5 41.1 41.6 42.6
Aa2 14.7 1.2 12.3 13.8 14.7 15.4 17.1
Bb2 4.7 1.2 2.3 3.9 4.7 5.5 7.1
Bb3 -0.7 1.2 -3.0 -1.5 -0.7 0.1 1.6
Aa2:Bb2 -15.7 1.7 -19.2 -16.8 -15.7 -14.7 -12.4
Aa2:Bb3 9.3 1.7 5.9 8.2 9.3 10.4 12.7
sigma 2.7 0.3 2.2 2.5 2.6 2.8 3.2
mean_PPD 48.7 0.5 47.7 48.3 48.7 49.0 49.6
log-posterior -158.5 2.0 -163.2 -159.5 -158.1 -157.0 -155.7
Diagnostics:
mcse Rhat n_eff
(Intercept) 0.0 1.0 1880
Aa2 0.0 1.0 1992
Bb2 0.0 1.0 1987
Bb3 0.0 1.0 2178
Aa2:Bb2 0.0 1.0 2150
Aa2:Bb3 0.0 1.0 2027
sigma 0.0 1.0 1543
mean_PPD 0.0 1.0 1778
log-posterior 0.1 1.0 1177
For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).
# OR library(broom) tidyMCMC(data.rstanarm$stanfit, conf.int = TRUE, conf.method = "HPDinterval", rhat = TRUE, ess = TRUE)
term estimate std.error conf.low conf.high rhat ess 1 (Intercept) 41.0695427 0.8462585 39.407984 42.715661 1.0008443 1880 2 Aa2 14.6591690 1.1981562 12.398261 17.112206 1.0009174 1992 3 Bb2 4.6819518 1.1857841 2.323231 6.980180 1.0016083 1987 4 Bb3 -0.7107484 1.1808212 -2.927747 1.642205 1.0000224 2178 5 Aa2:Bb2 -15.7289876 1.6946307 -19.022967 -12.286452 1.0019440 2150 6 Aa2:Bb3 9.2932738 1.7176328 6.132746 12.911395 1.0018266 2027 7 sigma 2.6549943 0.2653598 2.169464 3.191681 0.9997383 1543 8 mean_PPD 48.6588609 0.4962977 47.703797 49.608743 0.9994542 1778 9 log-posterior -158.4548359 1.9549368 -162.471066 -155.473098 0.9996055 1177
- the intercept represents the mean of the first combination Aa1:Bb1 is
41.0695427 - Aa2:Bb1 is
14.659169units greater than Aa1:Bb1 - Aa1:Bb2 is
4.6819518units greater Aa1:Bb1 - Aa1:Bb3 is
-0.7107484units greater Aa1:Bb1 - Aa2:Bb2 is
-15.7289876units greater than the difference between (Aa1:Bb2 + Aa2:Bb1) and (2*Aa1:Bb1) - Aa2:Bb3 is
9.2932738units greater than the difference between (Aa1:Bb3 + Aa2:Bb1) and (2*Aa1:Bb1)
While workers attempt to become comfortable with a new statistical framework, it is only natural that they like to evaluate and comprehend new structures and output alongside more familiar concepts. One way to facilitate this is via Bayesian p-values that are somewhat analogous to the frequentist p-values for investigating the hypothesis that a parameter is equal to zero.
## since values are less than zero mcmcpvalue(as.matrix(data.rstanarm)[, "Aa2"])
[1] 0
mcmcpvalue(as.matrix(data.rstanarm)[, "Bb2"])
[1] 0.0004444444
mcmcpvalue(as.matrix(data.rstanarm)[, "Bb3"])
[1] 0.5448889
mcmcpvalue(as.matrix(data.rstanarm)[, "Aa2:Bb2"])
[1] 0
mcmcpvalue(as.matrix(data.rstanarm)[, "Aa2:Bb3"])
[1] 0
mcmcpvalue(as.matrix(data.rstanarm)[, c("Aa2:Bb2", "Aa2:Bb3")])
[1] 0
There is evidence of an interaction between A and B.
library(loo) (full = loo(data.rstanarm))
Computed from 2250 by 60 log-likelihood matrix
Estimate SE
elpd_loo -146.8 6.1
p_loo 6.7 1.4
looic 293.5 12.2
All Pareto k estimates are good (k < 0.5)
See help('pareto-k-diagnostic') for details.
data.rstanarm.red = update(data.rstanarm, . ~ A + B)
Gradient evaluation took 7.9e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.79 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.183286 seconds (Warm-up)
0.127264 seconds (Sampling)
0.31055 seconds (Total)
Gradient evaluation took 1.7e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.102309 seconds (Warm-up)
0.143427 seconds (Sampling)
0.245736 seconds (Total)
Gradient evaluation took 1.7e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds.
Adjust your expectations accordingly!
Elapsed Time: 0.091882 seconds (Warm-up)
0.134913 seconds (Sampling)
0.226795 seconds (Total)
(reduced = loo(data.rstanarm.red))
Computed from 2250 by 60 log-likelihood matrix
Estimate SE
elpd_loo -194.8 3.6
p_loo 4.3 0.5
looic 389.7 7.2
All Pareto k estimates are good (k < 0.5)
See help('pareto-k-diagnostic') for details.
par(mfrow = 1:2, mar = c(5, 3.8, 1, 0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full, reduced)
elpd_diff se
-48.1 6.7
Matrix model (BRMS)
summary(data.brms)
Family: gaussian(identity)
Formula: y ~ A * B
Data: data (Number of observations: 60)
Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2;
total post-warmup samples = 2250
ICs: LOO = NA; WAIC = NA; R2 = NA
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
Intercept 41.12 0.85 39.44 42.92 1936 1
Aa2 14.63 1.20 12.27 16.87 1538 1
Bb2 4.62 1.20 2.29 7.01 1914 1
Bb3 -0.76 1.21 -3.13 1.58 2013 1
Aa2:Bb2 -15.68 1.71 -18.92 -12.25 1696 1
Aa2:Bb3 9.31 1.72 6.00 12.66 1713 1
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat
sigma 2.64 0.26 2.2 3.21 1401 1
Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
is a crude measure of effective sample size, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).
# OR library(broom) tidyMCMC(data.brms$fit, conf.int = TRUE, conf.method = "HPDinterval", rhat = TRUE, ess = TRUE)
term estimate std.error conf.low conf.high rhat ess 1 b_Intercept 41.1249801 0.8520689 39.282630 42.674204 0.9996522 1936 2 b_Aa2 14.6333534 1.1996304 12.239047 16.843446 1.0000296 1538 3 b_Bb2 4.6176257 1.2040485 2.334325 7.039437 0.9998751 1914 4 b_Bb3 -0.7612484 1.2100417 -3.033821 1.662261 1.0012004 2013 5 b_Aa2:Bb2 -15.6839628 1.7100995 -18.928721 -12.285494 0.9994896 1696 6 b_Aa2:Bb3 9.3101691 1.7191171 5.774495 12.433714 1.0022274 1713 7 sigma 2.6429857 0.2638466 2.158565 3.141588 1.0046579 1401
- the intercept represents the mean of the first combination Aa1:Bb1 is
41.1249801 - Aa2:Bb1 is
14.6333534units greater than Aa1:Bb1 - Aa1:Bb2 is
4.6176257units greater Aa1:Bb1 - Aa1:Bb3 is
-0.7612484units greater Aa1:Bb1 - Aa2:Bb2 is
-15.6839628units greater than the difference between (Aa1:Bb2 + Aa2:Bb1) and (2*Aa1:Bb1) - Aa2:Bb3 is
9.3101691units greater than the difference between (Aa1:Bb3 + Aa2:Bb1) and (2*Aa1:Bb1)
While workers attempt to become comfortable with a new statistical framework, it is only natural that they like to evaluate and comprehend new structures and output alongside more familiar concepts. One way to facilitate this is via Bayesian p-values that are somewhat analogous to the frequentist p-values for investigating the hypothesis that a parameter is equal to zero.
## since values are less than zero mcmcpvalue(as.matrix(data.brms)[, "b_Aa2"])
[1] 0
mcmcpvalue(as.matrix(data.brms)[, "b_Bb2"])
[1] 0
mcmcpvalue(as.matrix(data.brms)[, "b_Bb3"])
[1] 0.528
mcmcpvalue(as.matrix(data.brms)[, "b_Aa2:Bb2"])
[1] 0
mcmcpvalue(as.matrix(data.brms)[, "b_Aa2:Bb3"])
[1] 0
mcmcpvalue(as.matrix(data.brms)[, c("b_Aa2:Bb2", "b_Aa2:Bb3")])
[1] 0
There is evidence of an interaction between A and B.
library(loo) (full = loo(data.brms))
LOOIC SE 294.22 12.62
data.brms.red = update(data.brms, . ~ A + B)
SAMPLING FOR MODEL 'gaussian(identity) brms-model' NOW (CHAIN 1).
Gradient evaluation took 2.6e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.26 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 600 / 2000 [ 30%] (Warmup)
Iteration: 800 / 2000 [ 40%] (Warmup)
Iteration: 1000 / 2000 [ 50%] (Warmup)
Iteration: 1001 / 2000 [ 50%] (Sampling)
Iteration: 1200 / 2000 [ 60%] (Sampling)
Iteration: 1400 / 2000 [ 70%] (Sampling)
Iteration: 1600 / 2000 [ 80%] (Sampling)
Iteration: 1800 / 2000 [ 90%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.054481 seconds (Warm-up)
0.034883 seconds (Sampling)
0.089364 seconds (Total)
SAMPLING FOR MODEL 'gaussian(identity) brms-model' NOW (CHAIN 2).
Gradient evaluation took 1e-05 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 600 / 2000 [ 30%] (Warmup)
Iteration: 800 / 2000 [ 40%] (Warmup)
Iteration: 1000 / 2000 [ 50%] (Warmup)
Iteration: 1001 / 2000 [ 50%] (Sampling)
Iteration: 1200 / 2000 [ 60%] (Sampling)
Iteration: 1400 / 2000 [ 70%] (Sampling)
Iteration: 1600 / 2000 [ 80%] (Sampling)
Iteration: 1800 / 2000 [ 90%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.056183 seconds (Warm-up)
0.033942 seconds (Sampling)
0.090125 seconds (Total)
SAMPLING FOR MODEL 'gaussian(identity) brms-model' NOW (CHAIN 3).
Gradient evaluation took 7e-06 seconds
1000 transitions using 10 leapfrog steps per transition would take 0.07 seconds.
Adjust your expectations accordingly!
Iteration: 1 / 2000 [ 0%] (Warmup)
Iteration: 200 / 2000 [ 10%] (Warmup)
Iteration: 400 / 2000 [ 20%] (Warmup)
Iteration: 600 / 2000 [ 30%] (Warmup)
Iteration: 800 / 2000 [ 40%] (Warmup)
Iteration: 1000 / 2000 [ 50%] (Warmup)
Iteration: 1001 / 2000 [ 50%] (Sampling)
Iteration: 1200 / 2000 [ 60%] (Sampling)
Iteration: 1400 / 2000 [ 70%] (Sampling)
Iteration: 1600 / 2000 [ 80%] (Sampling)
Iteration: 1800 / 2000 [ 90%] (Sampling)
Iteration: 2000 / 2000 [100%] (Sampling)
Elapsed Time: 0.050325 seconds (Warm-up)
0.040119 seconds (Sampling)
0.090444 seconds (Total)
(reduced = loo(data.brms.red))
LOOIC SE 389.3 7.33
par(mfrow = 1:2, mar = c(5, 3.8, 1, 0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full, reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
Graphical summaries
A nice graphic is often a great accompaniment to a statistical analysis. Although there are no fixed assumptions associated with graphing (in contrast to statistical analyses), we often want the graphical summaries to reflect the associated statistical analyses. After all, the sample is just one perspective on the population(s). What we are more interested in is being able to estimate and depict likely population parameters/trends.
Thus, whilst we could easily provide a plot displaying the raw data along with simple measures of location and spread, arguably, we should use estimates that reflect the fitted model. In this case, it would be appropriate to plot the credibility interval associated with each group.
Matrix model (MCMCpack)
mcmc = data.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^A|^B", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(A=levels(data$A), B=levels(data$B)) Xmat = model.matrix(~A*B,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
A B estimate std.error conf.low conf.high 1 a1 b1 41.10322 0.8453382 39.47348 42.78445 2 a2 b1 55.74816 0.8396479 54.06831 57.38755 3 a1 b2 45.74120 0.8415863 44.05709 47.33058 4 a2 b2 44.67183 0.8364140 42.96477 46.25137 5 a1 b3 40.33618 0.8405389 38.67320 41.95839 6 a2 b3 64.33111 0.8360688 62.65830 65.92267
ggplot(newdata, aes(y=estimate, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

Matrix model (JAGS)
mcmc = data.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(A=levels(data$A), B=levels(data$B)) Xmat = model.matrix(~A*B,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
A B estimate std.error conf.low conf.high 1 a1 b1 41.09392 0.8444629 39.42483 42.72095 2 a2 b1 55.74079 0.8414034 54.09476 57.37608 3 a1 b2 45.74557 0.8461340 44.17292 47.50110 4 a2 b2 44.68068 0.8486124 42.99659 46.32094 5 a1 b3 40.34830 0.8424072 38.70237 42.02781 6 a2 b3 64.33167 0.8446976 62.62737 65.94871
ggplot(newdata, aes(y=estimate, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

Matrix model (STAN)
mcmc = as.matrix(data.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(A=levels(data$A), B=levels(data$B)) Xmat = model.matrix(~A*B,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
A B estimate std.error conf.low conf.high 1 a1 b1 41.12805 0.8374676 39.59698 42.79644 2 a2 b1 55.73285 0.8338807 54.12117 57.38026 3 a1 b2 45.72989 0.8467869 44.11524 47.46595 4 a2 b2 44.69504 0.8474554 42.96429 46.27580 5 a1 b3 40.33750 0.8404500 38.76233 41.95413 6 a2 b3 64.32720 0.8687497 62.71616 66.11319
ggplot(newdata, aes(y=estimate, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

Matrix model (RSTANARM)
## The simple way newdata = expand.grid(A=levels(data$A), B=levels(data$B)) fit = posterior_linpred(data.rstanarm, newdata = newdata) newdata = newdata %>% cbind(tidyMCMC(as.mcmc(fit),conf.int = TRUE,conf.method = "HPDinterval")) ggplot(newdata, aes(y=estimate, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

## Or from the posteriors mcmc = as.matrix(data.rstanarm) wch = c(which(colnames(mcmc)=='(Intercept)'), grep("^Aa|^Bb", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(A=levels(data$A), B=levels(data$B)) Xmat = model.matrix(~A*B,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
A B estimate std.error conf.low conf.high 1 a1 b1 41.06954 0.8462585 39.40798 42.71566 2 a2 b1 55.72871 0.8416961 54.02975 57.36504 3 a1 b2 45.75149 0.8263118 44.09889 47.31572 4 a2 b2 44.68168 0.8313474 42.95808 46.24587 5 a1 b3 40.35879 0.8410074 38.69045 41.99314 6 a2 b3 64.31124 0.8614734 62.67914 66.01741
ggplot(newdata, aes(y=estimate, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

Matrix model (BRMS)
## The simple way plot(marginal_effects(data.brms))



## OR eff=marginal_effects(data.brms) ggplot(eff[['A:B']], aes(y=estimate__, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=lower__, ymax=upper__))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

## Or from the posteriors mcmc = as.matrix(data.brms) wch = grep("^b_", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(A=levels(data$A), B=levels(data$B)) Xmat = model.matrix(~A*B,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
A B estimate std.error conf.low conf.high 1 a1 b1 41.12498 0.8520689 39.28263 42.67420 2 a2 b1 55.75833 0.8755420 53.96607 57.40839 3 a1 b2 45.74261 0.8424219 44.07701 47.37255 4 a2 b2 44.69200 0.8426376 42.99458 46.31306 5 a1 b3 40.36373 0.8468461 38.74868 42.06297 6 a2 b3 64.30725 0.8308393 62.73376 65.95684
ggplot(newdata, aes(y=estimate, x=B, fill=A)) + geom_blank() + geom_line(aes(x=as.numeric(B), linetype=A)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=A), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('B')+ scale_shape_manual('A',values=c(21,16))+ scale_fill_manual('A',values=c('white','black'))+ scale_linetype_manual('A',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(0,1), legend.position=c(0.05,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.size=unit(1,'cm'))

Finite Population Standard Deviations
Variance components, the amount of added variance attributed to each influence, are traditionally estimated for so called random effects. These are the effects for which the levels employed in the design are randomly selected to represent a broader range of possible levels. For such effects, effect sizes (differences between each level and a reference level) are of little value. Instead, the 'importance' of the variables are measured in units of variance components.
On the other hand, regular variance components for fixed factors (those whose measured levels represent the only levels of interest) are not logical - since variance components estimate variance as if the levels are randomly selected from a larger population. Nevertheless, in order to compare and contrast the scale of variability of both fixed and random factors, it is necessary to measure both on the same scale (sample or population based variance).
Finite-population variance components (Gelman, 2005) assume that the levels of all
factors (fixed and random) in the design are all the possible levels available.
In other words, they are assumed to represent finite populations of levels.
Sample (rather than population) statistics are then used to calculate these
finite-population variances (or standard deviations).
Since standard deviation (and variance) are bound at zero, standard deviation posteriors are typically non-normal. Consequently, medians and HPD intervals are more robust estimates.
library(broom) mcmc = data.mcmcpack Xmat = model.matrix(~A*B, data=data) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.A = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.B = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.AB = apply(cbind(0,mcmc[,which(wch %in% c(1,2,3))]),1,sd) # generate a model matrix newdata = data #expand.grid(A=levels(data$A), B=levels(data$B)) ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^Aa|^Bb',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,data$y,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.A, sd.B, sd.AB, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 10.355538 0.8468937 8.654072 11.974387 2 sd.B 2.985732 0.5877716 1.857284 4.146312 3 sd.AB 10.519220 0.7080347 9.091141 11.853109 4 sd.resid 2.602476 0.0762939 2.493373 2.750227
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 39.200390 1.7922327 35.666403 42.64867 2 sd.B 11.287484 1.7969726 7.535105 14.57798 3 sd.AB 39.772609 0.7968253 38.136506 41.25596 4 sd.resid 9.763594 0.7299524 8.669173 11.33111
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

Conclusions:
Approximately 39.8%
of the total finite population standard deviation is due to the interaction between factor A and factor B.
library(broom) mcmc = data.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~A*B, data=data) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.A = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.B = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.AB = apply(cbind(0,mcmc[,which(wch %in% c(1,2,3))]),1,sd) # generate a model matrix newdata = data #expand.grid(A=levels(data$A), B=levels(data$B)) ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,data$y,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.A, sd.B, sd.AB, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 10.356903 0.84402859 8.741790 12.049362 2 sd.B 2.985528 0.59407117 1.806780 4.142057 3 sd.AB 10.516470 0.70989309 9.154342 11.940921 4 sd.resid 2.603792 0.07735547 2.491510 2.753539
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 39.231736 1.7990769 35.637452 42.64830 2 sd.B 11.264562 1.8167232 7.733883 14.89522 3 sd.AB 39.768551 0.8060453 38.130117 41.29207 4 sd.resid 9.769675 0.7339516 8.659971 11.34210
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

Conclusions:
Approximately 39.8%
of the total finite population standard deviation is due to the interaction between factor A and factor B.
library(broom) mcmc = as.matrix(data.rstan) Xmat = model.matrix(~A*B, data=data) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.A = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.B = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.AB = apply(cbind(0,mcmc[,which(wch %in% c(1,2,3))]),1,sd) # generate a model matrix newdata = data #expand.grid(A=levels(data$A), B=levels(data$B)) ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,data$y,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.A, sd.B, sd.AB, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 10.327153 0.82874494 8.693664 11.919197 2 sd.B 2.971872 0.59809794 1.768250 4.106530 3 sd.AB 10.485393 0.69162110 9.190291 11.834249 4 sd.resid 2.603477 0.07920708 2.493581 2.759733
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 39.207844 1.7762236 35.304441 42.26207 2 sd.B 11.258603 1.8230887 7.233910 14.49477 3 sd.AB 39.776587 0.8382798 37.895241 41.19441 4 sd.resid 9.799974 0.7270559 8.707745 11.33364
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

Conclusions:
Approximately 39.8%
of the total finite population standard deviation is due to the interaction between factor A and factor B.
library(broom) mcmc = as.matrix(data.rstanarm) Xmat = model.matrix(~A*B, data=data) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.A = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.B = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.AB = apply(cbind(0,mcmc[,which(wch %in% c(1,2,3))]),1,sd) # generate a model matrix newdata = data #expand.grid(A=levels(data$A), B=levels(data$B)) ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^Aa|^Bb',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,data$y,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.A, sd.B, sd.AB, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 10.365598 0.84722438 8.766894 12.100157 2 sd.B 2.990030 0.58459570 1.828006 4.133714 3 sd.AB 10.519754 0.71389520 9.137064 11.948545 4 sd.resid 2.602193 0.07551823 2.492607 2.750141
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 39.194710 1.7532609 35.528072 42.47923 2 sd.B 11.281264 1.7835163 7.499541 14.65447 3 sd.AB 39.753790 0.7876428 38.208018 41.30859 4 sd.resid 9.771769 0.7275434 8.691907 11.36930
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

Conclusions:
Approximately 39.8%
of the total finite population standard deviation is due to the interaction between factor A and factor B.
library(broom) mcmc = as.matrix(data.brms) Xmat = model.matrix(~A*B, data=data) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.A = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.B = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.AB = apply(cbind(0,mcmc[,which(wch %in% c(1,2,3))]),1,sd) # generate a model matrix newdata = data #expand.grid(A=levels(data$A), B=levels(data$B)) ## get median parameter estimates wch = grep('^b_',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,data$y,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.A, sd.B, sd.AB, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 10.347343 0.84826682 8.654313 11.910115 2 sd.B 2.974928 0.58618754 1.908368 4.115285 3 sd.AB 10.501908 0.71427637 9.160508 11.878555 4 sd.resid 2.604827 0.08059903 2.490661 2.767553
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.A 39.286960 1.8237965 35.814048 42.86019 2 sd.B 11.214740 1.7806562 7.861384 14.69640 3 sd.AB 39.771178 0.8122531 38.202326 41.32874 4 sd.resid 9.779338 0.7431637 8.608327 11.36032
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

Conclusions:
Approximately 39.8%
of the total finite population standard deviation is due to the interaction between factor A and factor B.
$R^2$
In a frequentist context, the $R^2$ value is seen as a useful indicator of goodness of fit. Whilst it has long been acknowledged that this measure is not appropriate for comparing models (for such purposes information criterion such as AIC are more appropriate), it is nevertheless useful for estimating the amount (percent) of variance explained by the model.
In a frequentist context, $R^2$ is calculated as the variance in predicted values divided by the variance in the observed (response) values.
Unfortunately, this classical formulation does not translate simply into a Bayesian context since
the equivalently calculated numerator can be larger than the an equivalently calculated denominator - thereby resulting in an $R^2$
greater than 100%. Gelman, Goodrich, Gabry, and Ali (2017) proposed an alternative
formulation in which the denominator comprises the sum of the explained variance and the variance of the residuals.
So in the standard regression model notation of: $$ \begin{align} y_i \sim{}& N(\mu_i, \sigma)\\ \mu_i =& \mathbf{X}\boldsymbol{\beta} \end{align} $$ The $R^2$ could be formulated as: $$ R^2 = \frac{\sigma^2_f}{\sigma^2_f + \sigma^2_e} $$ where $\sigma^2_f = var(\mu)$, ($\mu = \mathbf{X}\boldsymbol{\beta})$) and for Gaussian models $\sigma^2_e = var(y-\mu)$
library(broom) mcmc <- data.mcmcpack Xmat = model.matrix(~A * B, data) wch = c(which(colnames(mcmc) == "(Intercept)"), grep("^Aa|^Bb", colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, data$y, "-") var_f = apply(fit, 1, var) var_e = apply(resid, 1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.9180017 0.007587991 0.903316 0.9290191
# for comparison with frequentist summary(lm(y ~ A * B, data))
Call:
lm(formula = y ~ A * B, data = data)
Residuals:
Min 1Q Median 3Q Max
-7.3944 -1.5753 0.2281 1.5575 5.1909
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 41.0988 0.8218 50.010 < 2e-16 ***
Aa2 14.6515 1.1622 12.606 < 2e-16 ***
Bb2 4.6386 1.1622 3.991 0.0002 ***
Bb3 -0.7522 1.1622 -0.647 0.5202
Aa2:Bb2 -15.7183 1.6436 -9.563 3.24e-13 ***
Aa2:Bb3 9.3352 1.6436 5.680 5.54e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.599 on 54 degrees of freedom
Multiple R-squared: 0.9245, Adjusted R-squared: 0.9175
F-statistic: 132.3 on 5 and 54 DF, p-value: < 2.2e-16
library(broom) mcmc <- data.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~A * B, data) wch = grep("^beta", colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, data$y, "-") var_f = apply(fit, 1, var) var_e = apply(resid, 1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.9178823 0.007656105 0.9026284 0.9291628
# for comparison with frequentist summary(lm(y ~ A * B, data))
Call:
lm(formula = y ~ A * B, data = data)
Residuals:
Min 1Q Median 3Q Max
-7.3944 -1.5753 0.2281 1.5575 5.1909
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 41.0988 0.8218 50.010 < 2e-16 ***
Aa2 14.6515 1.1622 12.606 < 2e-16 ***
Bb2 4.6386 1.1622 3.991 0.0002 ***
Bb3 -0.7522 1.1622 -0.647 0.5202
Aa2:Bb2 -15.7183 1.6436 -9.563 3.24e-13 ***
Aa2:Bb3 9.3352 1.6436 5.680 5.54e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.599 on 54 degrees of freedom
Multiple R-squared: 0.9245, Adjusted R-squared: 0.9175
F-statistic: 132.3 on 5 and 54 DF, p-value: < 2.2e-16
library(broom) mcmc <- as.matrix(data.rstan) Xmat = model.matrix(~A * B, data) wch = grep("^beta", colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, data$y, "-") var_f = apply(fit, 1, var) var_e = apply(resid, 1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.9177988 0.007611679 0.9039957 0.9288649
# for comparison with frequentist summary(lm(y ~ A * B, data))
Call:
lm(formula = y ~ A * B, data = data)
Residuals:
Min 1Q Median 3Q Max
-7.3944 -1.5753 0.2281 1.5575 5.1909
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 41.0988 0.8218 50.010 < 2e-16 ***
Aa2 14.6515 1.1622 12.606 < 2e-16 ***
Bb2 4.6386 1.1622 3.991 0.0002 ***
Bb3 -0.7522 1.1622 -0.647 0.5202
Aa2:Bb2 -15.7183 1.6436 -9.563 3.24e-13 ***
Aa2:Bb3 9.3352 1.6436 5.680 5.54e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.599 on 54 degrees of freedom
Multiple R-squared: 0.9245, Adjusted R-squared: 0.9175
F-statistic: 132.3 on 5 and 54 DF, p-value: < 2.2e-16
library(broom) mcmc <- as.matrix(data.rstanarm) Xmat = model.matrix(~A * B, data) wch = c(which(colnames(mcmc) == "(Intercept)"), grep("^Aa|^Bb", colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, data$y, "-") var_f = apply(fit, 1, var) var_e = apply(resid, 1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.917844 0.007763295 0.9038677 0.9290854
# for comparison with frequentist summary(lm(y ~ A * B, data))
Call:
lm(formula = y ~ A * B, data = data)
Residuals:
Min 1Q Median 3Q Max
-7.3944 -1.5753 0.2281 1.5575 5.1909
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 41.0988 0.8218 50.010 < 2e-16 ***
Aa2 14.6515 1.1622 12.606 < 2e-16 ***
Bb2 4.6386 1.1622 3.991 0.0002 ***
Bb3 -0.7522 1.1622 -0.647 0.5202
Aa2:Bb2 -15.7183 1.6436 -9.563 3.24e-13 ***
Aa2:Bb3 9.3352 1.6436 5.680 5.54e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.599 on 54 degrees of freedom
Multiple R-squared: 0.9245, Adjusted R-squared: 0.9175
F-statistic: 132.3 on 5 and 54 DF, p-value: < 2.2e-16
library(broom) mcmc <- as.matrix(data.brms) Xmat = model.matrix(~A * B, data) wch = grep("^b_", colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, data$y, "-") var_f = apply(fit, 1, var) var_e = apply(resid, 1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.9176029 0.00786278 0.9024319 0.9290937
# for comparison with frequentist summary(lm(y ~ A * B, data))
Call:
lm(formula = y ~ A * B, data = data)
Residuals:
Min 1Q Median 3Q Max
-7.3944 -1.5753 0.2281 1.5575 5.1909
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 41.0988 0.8218 50.010 < 2e-16 ***
Aa2 14.6515 1.1622 12.606 < 2e-16 ***
Bb2 4.6386 1.1622 3.991 0.0002 ***
Bb3 -0.7522 1.1622 -0.647 0.5202
Aa2:Bb2 -15.7183 1.6436 -9.563 3.24e-13 ***
Aa2:Bb3 9.3352 1.6436 5.680 5.54e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.599 on 54 degrees of freedom
Multiple R-squared: 0.9245, Adjusted R-squared: 0.9175
F-statistic: 132.3 on 5 and 54 DF, p-value: < 2.2e-16
Dealing with interactions
In the current working example, we have identified that there is a significant interaction between Factor A and Factor B. Our exploration of the regression coefficients, indicated that the pattern between b1, b2 and b3 might differ between a1 and a2.
Similarly, if we consider the coefficients from the perspective of Factor A, we can see that the patterns between a1 and a2 are similar for b1 and b3, yet very different for b2.
At this point, we can then split the two-factor model up into a series of single-factor models, either:
- examining the effects of Factor B separately for each level of Factor A (two single-factor models) or
- examining the effects of Factor A separately for each level of Factor B (three single-factor models)
However, rather than subset the data and fit isolated smaller models, it is arguably better to treat these explorations as contrasts. As such we could either:
- apply specific contrasts to the already fit model
- define the specific contrasts and use them to refit the model
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- data.mcmcpack wch = c(which(colnames(mcmc) == "(Intercept)"), grep("^Aa|^Bb", colnames(mcmc))) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) head(fit)
1 2 3 4 5 6 [1,] 40.42172 54.59308 44.36348 44.47316 40.09173 64.73362 [2,] 41.37944 56.20291 45.77572 43.86244 39.13803 63.79749 [3,] 40.90443 55.51086 44.90131 46.86885 38.79658 63.35621 [4,] 41.13543 56.87889 46.14153 43.72701 39.97623 65.45726 [5,] 41.09695 56.83938 45.91303 45.19079 41.35012 64.51791 [6,] 40.77523 55.82232 46.70431 45.13178 41.48085 64.93043
## we want to compare columns 2-1, 4-3 and 6-5 comps = fit[, c(2, 4, 6)] - fit[, c(1, 3, 5)] tidyMCMC(comps, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 2 14.644943 1.197689 12.238706 16.934341 2 4 -1.069371 1.186164 -3.610838 1.084133 3 6 23.994938 1.178228 21.626164 26.276557
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- data.mcmcpack wch = c(which(colnames(mcmc) == "(Intercept)"), grep("^Aa|^Bb", colnames(mcmc))) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) contr = attr(Xmat, "contrasts") newdata.a1 = model.frame(~A * B, expand.grid(A = levels(data$A)[1], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a1 = model.matrix(~A * B, data = newdata.a1, contrasts = contr) newdata.a2 = model.frame(~A * B, expand.grid(A = levels(data$A)[2], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a2 = model.matrix(~A * B, data = newdata.a2, contrasts = contr) Xmat = Xmat.a2 - Xmat.a1 coefs = mcmc[, wch] fit = coefs %*% t(Xmat) tidyMCMC(fit, conf.int = TRUE, conf.method = "HPDinterval")
estimate std.error conf.low conf.high 1 14.644943 1.197689 12.238706 16.934341 2 -1.069371 1.186164 -3.610838 1.084133 3 23.994938 1.178228 21.626164 26.276557
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- data.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) head(fit)
1 2 3 4 5 6 [1,] 40.48339 55.54703 46.41903 43.61135 40.17401 63.28148 [2,] 41.63781 56.00773 43.56967 44.24817 40.48599 64.59584 [3,] 40.61291 55.54098 45.24972 45.04578 40.11649 65.41953 [4,] 42.04693 56.55105 47.58341 44.47991 39.84335 63.02927 [5,] 42.38741 55.33531 46.50659 45.22567 41.49760 66.16237 [6,] 40.14774 55.55227 44.75628 44.08303 42.36898 63.97392
## we want to compare columns 2-1, 4-3 and 6-5 comps = fit[, c(2, 4, 6)] - fit[, c(1, 3, 5)] tidyMCMC(comps, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 2 14.646873 1.193637 12.362757 17.040371 2 4 -1.064884 1.201019 -3.373144 1.360905 3 6 23.983372 1.184993 21.704816 26.352716
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- data.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) contr = attr(Xmat, "contrasts") newdata.a1 = model.frame(~A * B, expand.grid(A = levels(data$A)[1], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a1 = model.matrix(~A * B, data = newdata.a1, contrasts = contr) newdata.a2 = model.frame(~A * B, expand.grid(A = levels(data$A)[2], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a2 = model.matrix(~A * B, data = newdata.a2, contrasts = contr) Xmat = Xmat.a2 - Xmat.a1 coefs = mcmc[, wch] fit = coefs %*% t(Xmat) tidyMCMC(fit, conf.int = TRUE, conf.method = "HPDinterval")
estimate std.error conf.low conf.high 1 14.646873 1.193637 12.362757 17.040371 2 -1.064884 1.201019 -3.373144 1.360905 3 23.983372 1.184993 21.704816 26.352716
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- as.matrix(data.rstan) wch = grep("^beta", colnames(mcmc)) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) head(fit)
iterations 1 2 3 4 5 6 [1,] 41.51135 55.70492 45.25656 44.02626 39.27705 64.13068 [2,] 42.17704 56.91515 44.11912 46.12462 39.82166 61.97899 [3,] 40.71477 55.71899 46.11131 44.60881 39.56221 65.73200 [4,] 40.81325 55.26740 45.44547 44.15927 39.35125 65.22235 [5,] 41.84887 56.16573 45.18476 45.40723 39.62631 62.76526 [6,] 41.89424 56.48574 45.96376 45.60291 39.74714 64.15358## we want to compare columns 2-1, 4-3 and 6-5 comps = fit[, c(2, 4, 6)] - fit[, c(1, 3, 5)] tidyMCMC(comps, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 2 14.604800 1.172022 12.294697 16.856289 2 4 -1.034849 1.168895 -3.295307 1.403327 3 6 23.989705 1.186497 21.769016 26.332830
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- as.matrix(data.rstan) wch = grep("^beta", colnames(mcmc)) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) contr = attr(Xmat, "contrasts") newdata.a1 = model.frame(~A * B, expand.grid(A = levels(data$A)[1], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a1 = model.matrix(~A * B, data = newdata.a1, contrasts = contr) newdata.a2 = model.frame(~A * B, expand.grid(A = levels(data$A)[2], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a2 = model.matrix(~A * B, data = newdata.a2, contrasts = contr) Xmat = Xmat.a2 - Xmat.a1 coefs = mcmc[, wch] fit = coefs %*% t(Xmat) tidyMCMC(fit, conf.int = TRUE, conf.method = "HPDinterval")
estimate std.error conf.low conf.high 1 14.604800 1.172022 12.294697 16.856289 2 -1.034849 1.168895 -3.295307 1.403327 3 23.989705 1.186497 21.769016 26.332830
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- as.matrix(data.rstanarm) wch = c(which(colnames(mcmc) == "(Intercept)"), grep("^Aa|^Bb", colnames(mcmc))) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) head(fit)
iterations 1 2 3 4 5 6 [1,] 41.69558 56.63831 46.05740 44.41792 39.69553 63.50611 [2,] 40.42830 54.97709 47.05839 45.21893 40.10380 64.94983 [3,] 42.20118 55.40128 45.66861 44.73203 40.65879 63.96776 [4,] 40.05899 56.39807 46.23925 44.72761 41.06437 63.17787 [5,] 39.79187 56.44716 46.91017 44.20343 40.52490 63.51040 [6,] 40.93069 55.74621 47.10475 43.89062 39.40017 65.50021## we want to compare columns 2-1, 4-3 and 6-5 comps = fit[, c(2, 4, 6)] - fit[, c(1, 3, 5)] tidyMCMC(comps, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 2 14.659169 1.198156 12.398261 17.112206 2 4 -1.069819 1.150765 -3.198597 1.284747 3 6 23.952443 1.198063 21.671556 26.284045
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- as.matrix(data.rstanarm) wch = c(which(colnames(mcmc) == "(Intercept)"), grep("^Aa|^Bb", colnames(mcmc))) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) contr = attr(Xmat, "contrasts") newdata.a1 = model.frame(~A * B, expand.grid(A = levels(data$A)[1], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a1 = model.matrix(~A * B, data = newdata.a1, contrasts = contr) newdata.a2 = model.frame(~A * B, expand.grid(A = levels(data$A)[2], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a2 = model.matrix(~A * B, data = newdata.a2, contrasts = contr) Xmat = Xmat.a2 - Xmat.a1 coefs = mcmc[, wch] fit = coefs %*% t(Xmat) tidyMCMC(fit, conf.int = TRUE, conf.method = "HPDinterval")
estimate std.error conf.low conf.high 1 14.659169 1.198156 12.398261 17.112206 2 -1.069819 1.150765 -3.198597 1.284747 3 23.952443 1.198063 21.671556 26.284045
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- as.matrix(data.brms) wch = grep("^b_", colnames(mcmc)) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) head(fit)
iterations 1 2 3 4 5 6 [1,] 43.16788 56.22417 46.04023 44.57059 40.20208 66.10624 [2,] 40.60622 57.95910 43.97139 44.50581 41.17112 63.10272 [3,] 40.28487 57.02149 46.15325 44.93627 39.85675 64.08042 [4,] 39.42845 56.34337 45.68384 43.00036 37.97875 64.22784 [5,] 41.34114 55.80752 46.66046 45.29682 39.28842 63.88255 [6,] 41.16159 56.16514 47.31944 46.47054 41.13258 64.86110## we want to compare columns 2-1, 4-3 and 6-5 comps = fit[, c(2, 4, 6)] - fit[, c(1, 3, 5)] tidyMCMC(comps, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 2 14.633353 1.199630 12.239047 16.843446 2 4 -1.050609 1.219472 -3.535571 1.198287 3 6 23.943522 1.212319 21.588699 26.249083
- by generating the posteriors of the cell means (means of each factor combination) and then
manually compare the appropriate columns for specific levels of factor B.
library(broom) mcmc <- as.matrix(data.brms) wch = grep("^b_", colnames(mcmc)) newdata = expand.grid(A = levels(data$A), B = levels(data$B)) Xmat = model.matrix(~A * B, data = newdata) contr = attr(Xmat, "contrasts") newdata.a1 = model.frame(~A * B, expand.grid(A = levels(data$A)[1], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a1 = model.matrix(~A * B, data = newdata.a1, contrasts = contr) newdata.a2 = model.frame(~A * B, expand.grid(A = levels(data$A)[2], B = levels(data$B)), xlev = list(A = levels(data$A), B = levels(data$B))) Xmat.a2 = model.matrix(~A * B, data = newdata.a2, contrasts = contr) Xmat = Xmat.a2 - Xmat.a1 coefs = mcmc[, wch] fit = coefs %*% t(Xmat) tidyMCMC(fit, conf.int = TRUE, conf.method = "HPDinterval")
estimate std.error conf.low conf.high 1 14.633353 1.199630 12.239047 16.843446 2 -1.050609 1.219472 -3.535571 1.198287 3 23.943522 1.212319 21.588699 26.249083
References
Gelman, A. (2005). “Analysis of Variance - Why it is More Important Than Ever”. In: The Annals of Statistics 33.1, pp. 1–53.
Gelman, A., B. Goodrich, J. Gabry, et al. (2017). “R-squared for Bayesian regression models”.
Worked Examples
- McCarthy (2007) - Chpt 6
- Kery (2010) - Chpt 10
- Gelman & Hill (2007) - Chpt 4
- Logan (2010) - Chpt 12
- Quinn & Keough (2002) - Chpt 9
Two-factor ANOVA
A biologist studying starlings wanted to know whether the mean mass of starlings differed according to different roosting situations. She was also interested in whether the mean mass of starlings altered over winter (Northern hemisphere) and whether the patterns amongst roosting situations were consistent throughout winter, therefore starlings were captured at the start (November) and end of winter (January). Ten starlings were captured from each roosting situation in each season, so in total, 80 birds were captured and weighed.
Download Starling data set| Format of starling.csv data files | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
starling <- read.table("../downloads/data/starling.csv", header = T, sep = ",", strip.white = T) head(starling)
SITUATION MONTH MASS GROUP 1 S1 November 78 S1Nov 2 S1 November 88 S1Nov 3 S1 November 87 S1Nov 4 S1 November 88 S1Nov 5 S1 November 83 S1Nov 6 S1 November 82 S1Nov
Exploratory data analysis did not reveal any issues with normality or homogeneity of variance.
- Fit the model to investigate the effects of situation and month on the
mass of starlings.
$$
\begin{align}
y_{ij} &\sim{} N(\mu_{ij}, \sigma)\\
\mu_{ij} &= \beta_0 + \mathbf{X}\boldsymbol{\beta}\\[1em]
\beta_0 &\sim{} N(0,100)\\
\beta &\sim{} N(0,10)\\
\sigma &\sim{} cauchy(0,5)\\
\end{align}
$$
library(MCMCpack) starling.mcmcpack = MCMCregress(MASS ~ SITUATION * MONTH, data = starling)
modelString = " model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(beta[],X[i,]) } #Priors beta0 ~ dnorm(0.01,1.0E-6) for (j in 1:nX) { beta[j] ~ dnorm(0.01,1.0E-6) } tau <- 1 / (sigma * sigma) sigma~dunif(0,100) } " X = model.matrix(~SITUATION * MONTH, data = starling) starling.list <- with(starling, list(y = MASS, X = X, nX = ncol(X), n = nrow(starling))) params <- c("beta", "sigma") burnInSteps = 3000 nChains = 3 numSavedSteps = 15000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) starling.r2jags <- jags(data = starling.list, inits = NULL, parameters.to.save = params, model.file = textConnection(modelString), n.chains = nChains, n.iter = nIter, n.burnin = burnInSteps, n.thin = thinSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph information: Observed stochastic nodes: 80 Unobserved stochastic nodes: 10 Total graph size: 844 Initializing model
modelString = " data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } transformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,100); sigma~cauchy(0,5); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " X = model.matrix(~SITUATION * MONTH, data = starling) starling.list <- with(starling, list(y = MASS, X = X, nX = ncol(X), n = nrow(starling))) starling.rstan <- stan(data = starling.list, model_code = modelString, chains = 3, iter = 2000, warmup = 500, thin = 3)
SAMPLING FOR MODEL '3d2414c9dcf4b5e12be870eadd2c894a' NOW (CHAIN 1). Gradient evaluation took 5.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.55 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.111089 seconds (Warm-up) 0.154646 seconds (Sampling) 0.265735 seconds (Total) SAMPLING FOR MODEL '3d2414c9dcf4b5e12be870eadd2c894a' NOW (CHAIN 2). Gradient evaluation took 9e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.100237 seconds (Warm-up) 0.16536 seconds (Sampling) 0.265597 seconds (Total) SAMPLING FOR MODEL '3d2414c9dcf4b5e12be870eadd2c894a' NOW (CHAIN 3). Gradient evaluation took 1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.09569 seconds (Warm-up) 0.140967 seconds (Sampling) 0.236657 seconds (Total)print(starling.rstan, par = c("beta", "sigma"))
Inference for Stan model: 3d2414c9dcf4b5e12be870eadd2c894a. 3 chains, each with iter=2000; warmup=500; thin=3; post-warmup draws per chain=500, total post-warmup draws=1500. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 90.80 0.05 1.35 88.22 89.93 90.76 91.70 93.40 864 1 beta[2] -0.60 0.06 1.89 -4.33 -1.88 -0.65 0.66 3.41 1090 1 beta[3] -2.57 0.06 1.97 -6.28 -3.94 -2.62 -1.30 1.59 953 1 beta[4] -6.61 0.06 1.95 -10.37 -7.87 -6.67 -5.29 -2.79 1045 1 beta[5] -7.21 0.06 1.90 -10.99 -8.51 -7.26 -5.95 -3.52 997 1 beta[6] -3.63 0.08 2.68 -9.05 -5.33 -3.64 -1.92 1.51 1156 1 beta[7] -2.41 0.09 2.75 -7.91 -4.19 -2.38 -0.65 3.16 1037 1 beta[8] -1.51 0.08 2.74 -7.12 -3.34 -1.51 0.22 4.06 1122 1 sigma 4.26 0.01 0.36 3.66 4.00 4.23 4.48 5.06 1452 1 Samples were drawn using NUTS(diag_e) at Sun Dec 17 10:33:10 2017. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).starling.rstanarm = stan_glm(MASS ~ SITUATION * MONTH, data = starling, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior_intercept = normal(0, 100), prior = normal(0, 100), prior_aux = cauchy(0, 2))
Gradient evaluation took 5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.5 seconds. Adjust your expectations accordingly! Elapsed Time: 0.145761 seconds (Warm-up) 0.312339 seconds (Sampling) 0.4581 seconds (Total) Gradient evaluation took 1.8e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.18 seconds. Adjust your expectations accordingly! Elapsed Time: 0.163082 seconds (Warm-up) 0.294581 seconds (Sampling) 0.457663 seconds (Total) Gradient evaluation took 1.9e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.19 seconds. Adjust your expectations accordingly! Elapsed Time: 0.155771 seconds (Warm-up) 0.292253 seconds (Sampling) 0.448024 seconds (Total)print(starling.rstanarm)
stan_glm family: gaussian [identity] formula: MASS ~ SITUATION * MONTH ------ Estimates: Median MAD_SD (Intercept) 90.8 1.3 SITUATIONS2 -0.6 1.9 SITUATIONS3 -2.6 1.9 SITUATIONS4 -6.7 1.9 MONTHNovember -7.2 1.9 SITUATIONS2:MONTHNovember -3.5 2.6 SITUATIONS3:MONTHNovember -2.3 2.7 SITUATIONS4:MONTHNovember -1.5 2.7 sigma 4.3 0.4 Sample avg. posterior predictive distribution of y (X = xbar): Median MAD_SD mean_PPD 83.8 0.7 ------ For info on the priors used see help('prior_summary.stanreg').tidyMCMC(starling.rstanarm, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 (Intercept) 90.8342507 1.3482297 88.384550 93.596736 2 SITUATIONS2 -0.6166141 1.9025010 -4.307238 3.045019 3 SITUATIONS3 -2.6053800 1.9154558 -6.321085 1.223459 4 SITUATIONS4 -6.6423833 1.9214525 -10.255593 -2.868400 5 MONTHNovember -7.2506064 1.8911235 -10.800146 -3.553678 6 SITUATIONS2:MONTHNovember -3.5640977 2.6910494 -8.963746 1.847141 7 SITUATIONS3:MONTHNovember -2.3913123 2.7221593 -7.680779 2.833313 8 SITUATIONS4:MONTHNovember -1.5670514 2.7163488 -6.709673 3.747383 9 sigma 4.2750066 0.3606119 3.617221 5.001696
starling.brms = brm(MASS ~ SITUATION * MONTH, data = starling, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior = c(prior(normal(0, 100), class = "Intercept"), prior(normal(0, 100), class = "b"), prior(cauchy(0, 5), class = "sigma")))
Gradient evaluation took 3.8e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.38 seconds. Adjust your expectations accordingly! Elapsed Time: 0.104651 seconds (Warm-up) 0.14288 seconds (Sampling) 0.247531 seconds (Total) Gradient evaluation took 1.1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds. Adjust your expectations accordingly! Elapsed Time: 0.105484 seconds (Warm-up) 0.149404 seconds (Sampling) 0.254888 seconds (Total) Gradient evaluation took 1.1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds. Adjust your expectations accordingly! Elapsed Time: 0.100582 seconds (Warm-up) 0.144617 seconds (Sampling) 0.245199 seconds (Total)print(starling.brms)
Family: gaussian(identity) Formula: MASS ~ SITUATION * MONTH Data: starling (Number of observations: 80) Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2; total post-warmup samples = 2250 ICs: LOO = NA; WAIC = NA; R2 = NA Population-Level Effects: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat Intercept 90.83 1.37 88.06 93.45 1533 1 SITUATIONS2 -0.64 1.97 -4.47 3.30 1573 1 SITUATIONS3 -2.64 1.91 -6.25 1.07 1455 1 SITUATIONS4 -6.62 1.95 -10.64 -2.81 1685 1 MONTHNovember -7.33 1.95 -11.25 -3.51 1503 1 SITUATIONS2:MONTHNovember -3.43 2.75 -8.92 1.89 1584 1 SITUATIONS3:MONTHNovember -2.21 2.68 -7.48 3.00 1579 1 SITUATIONS4:MONTHNovember -1.45 2.74 -6.79 4.08 1758 1 Family Specific Parameters: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat sigma 4.26 0.38 3.61 5.08 2026 1 Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat = 1).tidyMCMC(starling.brms, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 b_Intercept 90.8270337 1.3658808 88.241978 93.616386 2 b_SITUATIONS2 -0.6438853 1.9659375 -4.899954 2.814581 3 b_SITUATIONS3 -2.6449981 1.9064693 -6.267860 1.020327 4 b_SITUATIONS4 -6.6181945 1.9474139 -10.445554 -2.692970 5 b_MONTHNovember -7.3288521 1.9504259 -11.044855 -3.390492 6 b_SITUATIONS2:MONTHNovember -3.4343794 2.7496251 -9.012966 1.772736 7 b_SITUATIONS3:MONTHNovember -2.2091803 2.6833877 -7.424543 3.064406 8 b_SITUATIONS4:MONTHNovember -1.4513879 2.7449835 -6.722984 4.129929 9 sigma 4.2629104 0.3765409 3.522005 4.975503
- Explore MCMC diagnostics
library(MCMCpack) plot(starling.mcmcpack)



raftery.diag(starling.mcmcpack)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) (Intercept) 2 3802 3746 1.010 SITUATIONS2 1 3727 3746 0.995 SITUATIONS3 2 3788 3746 1.010 SITUATIONS4 2 3802 3746 1.010 MONTHNovember 2 3929 3746 1.050 SITUATIONS2:MONTHNovember 2 3771 3746 1.010 SITUATIONS3:MONTHNovember 2 3741 3746 0.999 SITUATIONS4:MONTHNovember 2 3710 3746 0.990 sigma2 2 3802 3746 1.010autocorr.diag(starling.mcmcpack)
(Intercept) SITUATIONS2 SITUATIONS3 SITUATIONS4 MONTHNovember SITUATIONS2:MONTHNovember Lag 0 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 Lag 1 -0.020582606 -0.027966067 -0.008357537 -0.008847059 -0.009698479 -0.027312974 Lag 5 -0.008067665 -0.003051190 -0.002691984 -0.002567663 0.001851634 -0.012010879 Lag 10 0.008907848 0.002499721 0.009703737 -0.009748247 -0.009797442 -0.005470297 Lag 50 0.012034643 -0.005892326 0.021304099 0.001993598 0.028984464 0.017210812 SITUATIONS3:MONTHNovember SITUATIONS4:MONTHNovember sigma2 Lag 0 1.000000000 1.000000000 1.000000000 Lag 1 0.004697856 -0.012495936 0.087368555 Lag 5 0.009598076 -0.009052204 -0.020665842 Lag 10 -0.007422252 -0.006324498 -0.002938015 Lag 50 0.012401999 0.015822469 0.009494986starling.mcmc = as.mcmc(starling.r2jags) plot(starling.mcmc)




preds <- grep("beta", colnames(starling.mcmc[[1]])) plot(starling.mcmc[, preds])


raftery.diag(starling.mcmc)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 35750 3746 9.54 beta[2] 20 36380 3746 9.71 beta[3] 20 38330 3746 10.20 beta[4] 10 37660 3746 10.10 beta[5] 10 37660 3746 10.10 beta[6] 20 38330 3746 10.20 beta[7] 20 37020 3746 9.88 beta[8] 20 39680 3746 10.60 deviance 20 37020 3746 9.88 sigma 20 37020 3746 9.88 [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 35750 3746 9.54 beta[2] 20 36380 3746 9.71 beta[3] 20 38330 3746 10.20 beta[4] 20 39000 3746 10.40 beta[5] 20 38330 3746 10.20 beta[6] 20 39000 3746 10.40 beta[7] 10 37660 3746 10.10 beta[8] 20 39000 3746 10.40 deviance 20 37020 3746 9.88 sigma 20 37020 3746 9.88 [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 10 37660 3746 10.10 beta[2] 20 37020 3746 9.88 beta[3] 20 36380 3746 9.71 beta[4] 20 39000 3746 10.40 beta[5] 10 37660 3746 10.10 beta[6] 20 38330 3746 10.20 beta[7] 10 37660 3746 10.10 beta[8] 20 37020 3746 9.88 deviance 10 37660 3746 10.10 sigma 20 35750 3746 9.54autocorr.diag(starling.mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] Lag 0 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 Lag 10 0.020018454 0.008385600 0.006773136 0.016767372 0.001794959 0.002023408 -0.005424213 Lag 50 -0.002226832 -0.005022803 -0.004897598 -0.006928097 -0.004534439 -0.003263828 -0.010696208 Lag 100 0.002965946 0.011904882 0.011257944 -0.009425998 0.006400201 0.012428418 0.011962211 Lag 500 -0.016334707 0.001892880 -0.019613370 -0.001546560 -0.015209895 -0.014686469 -0.019997432 beta[8] deviance sigma Lag 0 1.0000000000 1.000000000 1.000000000 Lag 10 0.0081625054 -0.003570058 -0.007876734 Lag 50 -0.0096533429 -0.003112686 0.006055349 Lag 100 -0.0007394014 0.005563932 -0.001960111 Lag 500 -0.0091243941 0.012633741 -0.001078114s = as.array(starling.rstan) wch = grep("beta", dimnames(s)$parameters) s = s[, , wch] mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)


autocorr.diag(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] Lag 0 1.00000000 1.000000000 1.000000000 1.00000000 1.000000000 1.00000000 1.00000000 Lag 1 0.16945363 0.109737457 0.150402279 0.10645605 0.122107238 0.06194732 0.12399164 Lag 5 0.03221219 0.015781535 0.018110384 -0.01565517 0.025473996 0.01971320 0.02429010 Lag 10 -0.02748358 0.003413902 -0.030686452 -0.01145101 -0.007227615 0.02094509 0.01470017 Lag 50 0.00762950 -0.002950695 -0.005933959 0.01183242 -0.023749263 -0.01424603 -0.02996707
## Or via rstan stan_trace(starling.rstan)

stan_ac(starling.rstan)

stan_rhat(starling.rstan)

stan_ess(starling.rstan)

## Or via bayesplot detach("package:reshape") mcmc_trace(as.matrix(starling.rstan), regex_pars = "beta|sigma")

mcmc_dens(as.matrix(starling.rstan), regex_pars = "beta|sigma")

s = as.array(starling.rstanarm) mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)


autocorr.diag(mcmc)
(Intercept) SITUATIONS2 SITUATIONS3 SITUATIONS4 MONTHNovember SITUATIONS2:MONTHNovember Lag 0 1.00000000 1.00000000 1.00000000 1.000000000 1.00000000 1.00000000 Lag 1 0.11159965 0.10573479 0.04719504 0.070848483 0.11479350 0.08716859 Lag 5 -0.01476018 -0.02549344 -0.03214101 0.011726487 -0.02662856 -0.02902201 Lag 10 0.01417466 -0.00921212 0.01760432 -0.005469897 -0.02261881 -0.02147699 Lag 50 0.01868028 0.04249583 0.04793095 0.002594378 0.01342523 0.02239605 SITUATIONS3:MONTHNovember SITUATIONS4:MONTHNovember Lag 0 1.000000000 1.000000000 Lag 1 0.076712991 0.096987790 Lag 5 -0.001611613 -0.012604740 Lag 10 0.004631691 -0.021466562 Lag 50 0.022316456 0.001137837## OR via rstan stan_trace(starling.rstanarm)

raftery.diag(starling.rstanarm)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
stan_ac(starling.rstanarm)

stan_rhat(starling.rstanarm)

stan_ess(starling.rstanarm)

## OR via bayesplot detach("package:reshape") mcmc_trace(as.array(starling.rstanarm), regex_pars = "Intercept|x|sigma")

mcmc_dens(as.array(starling.rstanarm))

posterior_vs_prior(starling.rstanarm, color_by = "vs", group_by = TRUE, facet_args = list(scales = "free_y"))
Gradient evaluation took 4.3e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.43 seconds. Adjust your expectations accordingly! Elapsed Time: 0.248229 seconds (Warm-up) 0.187957 seconds (Sampling) 0.436186 seconds (Total) Gradient evaluation took 1.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds. Adjust your expectations accordingly! Elapsed Time: 0.216608 seconds (Warm-up) 0.122935 seconds (Sampling) 0.339543 seconds (Total)
mcmc = as.mcmc(starling.brms) plot(mcmc)




autocorr.diag(mcmc)
Error in ts(x, start = start(x), end = end(x), deltat = thin(x)): invalid time series parameters specified
## OR via rstan stan_trace(starling.brms$fit)

raftery.diag(starling.brms)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
stan_ac(starling.brms$fit)

stan_rhat(starling.brms$fit)

stan_ess(starling.brms$fit)

- Explore model validation
mcmc = as.data.frame(starling.mcmcpack) #generate a model matrix newdata = starling Xmat = model.matrix(~SITUATION*MONTH, newdata) ##get median parameter estimates head(mcmc)
(Intercept) SITUATIONS2 SITUATIONS3 SITUATIONS4 MONTHNovember SITUATIONS2:MONTHNovember 1 93.41320 -5.6342588 -8.5410083 -10.065326 -6.671472 -1.923439 2 90.12052 0.2776593 -0.7887616 -7.601482 -5.595692 -4.540829 3 90.44095 -2.8284137 -2.7824024 -6.734315 -5.998972 -2.876719 4 88.70704 -0.2954951 -0.5593803 -8.284445 -4.320155 -6.141591 5 91.59384 -1.9203988 -3.6010898 -6.568399 -7.277158 -3.818587 6 88.86650 1.5991905 -1.7601855 -4.509152 -5.005089 -5.924305 SITUATIONS3:MONTHNovember SITUATIONS4:MONTHNovember sigma2 1 -2.048904 -0.4526515 22.09981 2 -3.626985 -1.2928752 27.84430 3 -2.495392 -2.7297938 17.87759 4 -2.132988 -0.3882854 23.31116 5 -2.646437 -0.1454131 17.23485 6 -2.950157 -2.5860752 15.31409
wch = grepl('sigma2',colnames(mcmc))==0 coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = starling$MASS - fit ggplot() + geom_point(starling=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=SITUATION))

ggplot(newdata) + geom_point(aes(y=resid, x=MONTH))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model wch = grepl('sigma',colnames(mcmc))==0 coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~SITUATION*MONTH, data=starling) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(starling), fit[i,], sqrt(mcmc[i, 'sigma2']))) newdata = data.frame(SITUATION=starling$SITUATION, MONTH=starling$MONTH, yRep) %>% gather(key=Sample, value=Value,-SITUATION,-MONTH) ggplot(newdata) + geom_violin(aes(y=Value, x=SITUATION, color=MONTH,fill='Model'), alpha=0.5)+ geom_violin(data=starling, aes(y=MASS,x=SITUATION,color=MONTH,fill='Obs'), alpha=0.5) + geom_point(data=starling, aes(y=MASS, x=SITUATION, color=MONTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=MONTH, fill='Model', group=MONTH, color=SITUATION), alpha=0.5)+ geom_point(data=starling, aes(y=MASS, x=MONTH, group=MONTH,color=SITUATION))

mcmc_intervals(as.matrix(starling.mcmcpack), regex_pars='Intercept|^SITUATION|^MONTH|sigma')

mcmc_areas(as.matrix(starling.mcmcpack), regex_pars='Intercept|^SITUATION|^MONTH|sigma')

mcmc = starling.r2jags$BUGSoutput$sims.matrix #generate a model matrix newdata = starling Xmat = model.matrix(~SITUATION*MONTH, newdata) ##get median parameter estimates wch = grep('^beta\\[',colnames(mcmc)) coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = starling$MASS - fit ggplot() + geom_point(starling=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=SITUATION))

ggplot(newdata) + geom_point(aes(y=resid, x=MONTH))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~SITUATION*MONTH, data=starling) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(starling), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(SITUATION=starling$SITUATION, MONTH=starling$MONTH, yRep) %>% gather(key=Sample, value=Value,-SITUATION,-MONTH) ggplot(newdata) + geom_violin(aes(y=Value, x=SITUATION, color=MONTH,fill='Model'), alpha=0.5)+ geom_violin(data=starling, aes(y=MASS,x=SITUATION,color=MONTH,fill='Obs'), alpha=0.5) + geom_point(data=starling, aes(y=MASS, x=SITUATION, color=MONTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=MONTH, fill='Model', group=MONTH, color=SITUATION), alpha=0.5)+ geom_point(data=starling, aes(y=MASS, x=MONTH, group=MONTH,color=SITUATION))

mcmc_intervals(starling.r2jags$BUGSoutput$sims.matrix, regex_pars='beta|sigma')

mcmc_areas(starling.r2jags$BUGSoutput$sims.matrix, regex_pars='beta|sigma')

mcmc = as.matrix(starling.rstan) #generate a model matrix newdata = starling Xmat = model.matrix(~SITUATION*MONTH, newdata) ##get median parameter estimates wch = grep('^beta\\[',colnames(mcmc)) coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = starling$MASS - fit ggplot() + geom_point(starling=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=SITUATION))

ggplot(newdata) + geom_point(aes(y=resid, x=MONTH))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~SITUATION*MONTH, data=starling) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(starling), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(SITUATION=starling$SITUATION, MONTH=starling$MONTH, yRep) %>% gather(key=Sample, value=Value,-SITUATION,-MONTH) ggplot(newdata) + geom_violin(aes(y=Value, x=SITUATION, color=MONTH,fill='Model'), alpha=0.5)+ geom_violin(data=starling, aes(y=MASS,x=SITUATION,color=MONTH,fill='Obs'), alpha=0.5) + geom_point(data=starling, aes(y=MASS, x=SITUATION, color=MONTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=MONTH, fill='Model', group=MONTH, color=SITUATION), alpha=0.5)+ geom_point(data=starling, aes(y=MASS, x=MONTH, group=MONTH,color=SITUATION))

mcmc_intervals(as.matrix(starling.rstan), regex_pars='^beta|sigma')

mcmc_areas(as.matrix(starling.rstan), regex_pars='^beta|sigma')

resid = resid(starling.rstanarm) fit = fitted(starling.rstanarm) ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

resid = resid(starling.rstanarm) dat = starling %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = SITUATION))

ggplot(dat) + geom_point(aes(y = resid, x = MONTH))

resid = resid(starling.rstanarm) sigma(starling.rstanarm)
[1] 4.25097
sresid = resid/sigma(starling.rstanarm) fit = fitted(starling.rstanarm) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

y_pred = posterior_predict(starling.rstanarm) newdata = starling %>% dplyr:::select(-GROUP) %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -SITUATION,-MONTH,-MASS) head(newdata)
SITUATION MONTH MASS Rep Value 1 S1 November 78 1 83.61995 2 S1 November 88 1 80.98251 3 S1 November 87 1 84.48206 4 S1 November 88 1 86.02628 5 S1 November 83 1 86.32812 6 S1 November 82 1 92.79887
ggplot(newdata) + geom_violin(aes(y=Value, x=SITUATION, color=MONTH,fill='Model'), alpha=0.5)+ geom_violin(data=starling, aes(y=MASS,x=SITUATION,color=MONTH,fill='Obs'), alpha=0.5) + geom_point(data=starling, aes(y=MASS, x=SITUATION, color=MONTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=MONTH, fill='Model', group=MONTH, color=SITUATION), alpha=0.5)+ geom_point(data=starling, aes(y=MASS, x=MONTH, group=MONTH,color=SITUATION))

mcmc_intervals(as.matrix(starling.rstanarm), regex_pars='Intercept|^SITUATION|^MONTH|sigma')

mcmc_areas(as.matrix(starling.rstanarm), regex_pars='Intercept|^SITUATION|^MONTH|sigma')

resid = resid(starling.brms)[,'Estimate'] fit = fitted(starling.brms)[,'Estimate'] ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

resid = resid(starling.brms)[,'Estimate'] dat = starling %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = SITUATION))

ggplot(dat) + geom_point(aes(y = resid, x = MONTH))

resid = resid(starling.brms) sresid = resid(starling.brms, type='pearson')[,'Estimate'] fit = fitted(starling.brms)[,'Estimate'] ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

y_pred = posterior_predict(starling.brms) newdata = starling %>% dplyr:::select(-GROUP) %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -SITUATION,-MONTH,-MASS) head(newdata)
SITUATION MONTH MASS Rep Value 1 S1 November 78 1 82.70407 2 S1 November 88 1 82.37511 3 S1 November 87 1 72.91501 4 S1 November 88 1 83.16775 5 S1 November 83 1 80.04324 6 S1 November 82 1 83.56222
ggplot(newdata) + geom_violin(aes(y=Value, x=SITUATION, color=MONTH,fill='Model'), alpha=0.5)+ geom_violin(data=starling, aes(y=MASS,x=SITUATION,color=MONTH,fill='Obs'), alpha=0.5) + geom_point(data=starling, aes(y=MASS, x=SITUATION, color=MONTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=MONTH, fill='Model', group=MONTH, color=SITUATION), alpha=0.5)+ geom_point(data=starling, aes(y=MASS, x=MONTH, group=MONTH,color=SITUATION))

mcmc_intervals(as.matrix(starling.brms), regex_pars='^b_|sigma')

mcmc_areas(as.matrix(starling.brms), regex_pars='^b_|sigma')
 All diagnostics seem reasonable.
All diagnostics seem reasonable.
- Explore parameter estimates
summary(starling.mcmcpack)
Iterations = 1001:11000 Thinning interval = 1 Number of chains = 1 Sample size per chain = 10000 1. Empirical mean and standard deviation for each variable, plus standard error of the mean: Mean SD Naive SE Time-series SE (Intercept) 90.7921 1.356 0.01356 0.01329 SITUATIONS2 -0.6032 1.931 0.01931 0.01878 SITUATIONS3 -2.5908 1.908 0.01908 0.02015 SITUATIONS4 -6.6016 1.907 0.01907 0.01915 MONTHNovember -7.2033 1.893 0.01893 0.01893 SITUATIONS2:MONTHNovember -3.5985 2.711 0.02711 0.02580 SITUATIONS3:MONTHNovember -2.4046 2.689 0.02689 0.02689 SITUATIONS4:MONTHNovember -1.5716 2.682 0.02682 0.02682 sigma2 18.1521 3.096 0.03096 0.03380 2. Quantiles for each variable: 2.5% 25% 50% 75% 97.5% (Intercept) 88.175 89.882 90.7828 91.6750 93.483 SITUATIONS2 -4.420 -1.894 -0.5911 0.6831 3.181 SITUATIONS3 -6.355 -3.847 -2.5872 -1.3177 1.094 SITUATIONS4 -10.285 -7.862 -6.6199 -5.3372 -2.864 MONTHNovember -10.945 -8.445 -7.1961 -5.9496 -3.478 SITUATIONS2:MONTHNovember -9.016 -5.365 -3.6086 -1.7990 1.720 SITUATIONS3:MONTHNovember -7.675 -4.195 -2.4052 -0.5881 2.842 SITUATIONS4:MONTHNovember -6.799 -3.379 -1.5691 0.2346 3.625 sigma2 13.128 15.924 17.7911 20.0008 25.095#OR library(broom) tidyMCMC(starling.mcmcpack,conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 (Intercept) 90.7921498 1.356498 88.211017 93.498231 2 SITUATIONS2 -0.6032381 1.931333 -4.516064 3.061980 3 SITUATIONS3 -2.5907839 1.908266 -6.273937 1.169130 4 SITUATIONS4 -6.6015829 1.907277 -10.284640 -2.857007 5 MONTHNovember -7.2032595 1.892917 -10.929176 -3.462664 6 SITUATIONS2:MONTHNovember -3.5984840 2.711370 -8.974449 1.743408 7 SITUATIONS3:MONTHNovember -2.4045698 2.688954 -7.694184 2.817359 8 SITUATIONS4:MONTHNovember -1.5715791 2.682241 -6.814495 3.604155 9 sigma2 18.1520730 3.096016 12.507476 24.203264
#OR with p-values newdata = with(starling, expand.grid(SITUATION=levels(SITUATION), MONTH=levels(MONTH))) Xmat = model.matrix(~SITUATION*MONTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(starling.mcmcpack[,i]) )
[1] 0 [1] 0.7554 [1] 0.1777 [1] 0.001 [1] 2e-04 [1] 0.1832 [1] 0.366 [1] 0.5588
# Main effect of SITUATION mcmcpvalue(starling.mcmcpack[,which(wch==1)])
[1] 0.0029
# Main effect of Month mcmcpvalue(starling.mcmcpack[,which(wch==2)])
[1] 2e-04
# Interaction mcmcpvalue(starling.mcmcpack[,which(wch==3)])
[1] 0.5892
## frequentist for comparison summary(lm(MASS~SITUATION*MONTH, data=starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14anova(lm(MASS~SITUATION*MONTH, data=starling))
Analysis of Variance Table Response: MASS Df Sum Sq Mean Sq F value Pr(>F) SITUATION 3 574.4 191.47 10.8207 5.960e-06 *** MONTH 1 1656.2 1656.20 93.6000 1.172e-14 *** SITUATION:MONTH 3 34.2 11.40 0.6443 0.5891 Residuals 72 1274.0 17.69 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1print(starling.r2jags)
Inference for Bugs model at "5", fit using jags, 3 chains, each with 50000 iterations (first 3000 discarded), n.thin = 10 n.sims = 14100 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 90.789 1.358 88.119 89.884 90.791 91.696 93.429 1.001 14000 beta[2] -0.589 1.917 -4.337 -1.887 -0.600 0.677 3.241 1.001 14000 beta[3] -2.598 1.913 -6.348 -3.865 -2.608 -1.332 1.203 1.001 14000 beta[4] -6.578 1.919 -10.334 -7.866 -6.592 -5.299 -2.772 1.001 14000 beta[5] -7.196 1.917 -10.942 -8.490 -7.210 -5.917 -3.422 1.001 14000 beta[6] -3.601 2.722 -8.970 -5.419 -3.603 -1.818 1.800 1.001 14000 beta[7] -2.404 2.716 -7.800 -4.197 -2.399 -0.626 2.890 1.001 11000 beta[8] -1.615 2.719 -6.949 -3.436 -1.610 0.230 3.718 1.001 6400 sigma 4.281 0.369 3.625 4.026 4.256 4.509 5.076 1.001 8000 deviance 458.129 4.568 451.263 454.837 457.454 460.652 469.101 1.001 14000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 10.4 and DIC = 468.6 DIC is an estimate of expected predictive error (lower deviance is better).#OR library(broom) tidyMCMC(starling.r2jags,conf.int=TRUE, conf.method='HPDinterval')
Error in colMeans(ss): 'x' must be numeric
#OR with p-values newdata = with(starling, expand.grid(SITUATION=levels(SITUATION), MONTH=levels(MONTH))) Xmat = model.matrix(~SITUATION*MONTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(starling.r2jags$BUGSoutput$sims.matrix[,i]) )
[1] 0 [1] 0.7576596 [1] 0.1724823 [1] 0.0007801418 [1] 0.0002836879 [1] 0.1848227 [1] 0.3687234 [1] 0.5521277
# Main effect of SITUATION mcmcpvalue(starling.r2jags$BUGSoutput$sims.matrix[,which(wch==1)])
[1] 0.003687943
# Main effect of Month mcmcpvalue(starling.r2jags$BUGSoutput$sims.matrix[,which(wch==2)])
[1] 0.0002836879
# Interaction mcmcpvalue(starling.r2jags$BUGSoutput$sims.matrix[,which(wch==3)])
[1] 0.5971631
## frequentist for comparison summary(lm(MASS~SITUATION*MONTH, data=starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14anova(lm(MASS~SITUATION*MONTH, data=starling))
Analysis of Variance Table Response: MASS Df Sum Sq Mean Sq F value Pr(>F) SITUATION 3 574.4 191.47 10.8207 5.960e-06 *** MONTH 1 1656.2 1656.20 93.6000 1.172e-14 *** SITUATION:MONTH 3 34.2 11.40 0.6443 0.5891 Residuals 72 1274.0 17.69 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1print(starling.rstan, pars=c('beta','sigma'))
Inference for Stan model: 3d2414c9dcf4b5e12be870eadd2c894a. 3 chains, each with iter=2000; warmup=500; thin=3; post-warmup draws per chain=500, total post-warmup draws=1500. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 90.80 0.05 1.35 88.22 89.93 90.76 91.70 93.40 864 1 beta[2] -0.60 0.06 1.89 -4.33 -1.88 -0.65 0.66 3.41 1090 1 beta[3] -2.57 0.06 1.97 -6.28 -3.94 -2.62 -1.30 1.59 953 1 beta[4] -6.61 0.06 1.95 -10.37 -7.87 -6.67 -5.29 -2.79 1045 1 beta[5] -7.21 0.06 1.90 -10.99 -8.51 -7.26 -5.95 -3.52 997 1 beta[6] -3.63 0.08 2.68 -9.05 -5.33 -3.64 -1.92 1.51 1156 1 beta[7] -2.41 0.09 2.75 -7.91 -4.19 -2.38 -0.65 3.16 1037 1 beta[8] -1.51 0.08 2.74 -7.12 -3.34 -1.51 0.22 4.06 1122 1 sigma 4.26 0.01 0.36 3.66 4.00 4.23 4.48 5.06 1452 1 Samples were drawn using NUTS(diag_e) at Sun Dec 17 10:33:10 2017. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).#OR library(broom) tidyMCMC(starling.rstan,conf.int=TRUE, conf.method='HPDinterval', pars=c('beta','sigma'))
term estimate std.error conf.low conf.high 1 beta[1] 90.8007947 1.3506028 88.160967 93.335626 2 beta[2] -0.6021332 1.8890495 -3.640324 3.844746 3 beta[3] -2.5656962 1.9697018 -6.524464 1.117853 4 beta[4] -6.6124813 1.9546114 -10.497149 -2.980277 5 beta[5] -7.2148225 1.8975842 -11.032805 -3.584344 6 beta[6] -3.6264954 2.6831763 -9.065828 1.520459 7 beta[7] -2.4119402 2.7536621 -8.225599 2.603148 8 beta[8] -1.5078933 2.7414112 -6.798526 4.323528 9 sigma 4.2625022 0.3566923 3.567918 4.931771
#OR with p-values newdata = with(starling, expand.grid(SITUATION=levels(SITUATION), MONTH=levels(MONTH))) Xmat = model.matrix(~SITUATION*MONTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(starling.rstan)[,i]) )
[1] 0 [1] 0.738 [1] 0.1746667 [1] 0.002 [1] 0 [1] 0.1746667 [1] 0.3673333 [1] 0.5653333
# Main effect of SITUATION mcmcpvalue(as.matrix(starling.rstan)[,which(wch==1)])
[1] 0.006666667
# Main effect of Month mcmcpvalue(as.matrix(starling.rstan)[,which(wch==2)])
[1] 0
# Interaction mcmcpvalue(as.matrix(starling.rstan)[,which(wch==3)])
[1] 0.56
## frequentist for comparison summary(lm(MASS~SITUATION*MONTH, data=starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14anova(lm(MASS~SITUATION*MONTH, data=starling))
Analysis of Variance Table Response: MASS Df Sum Sq Mean Sq F value Pr(>F) SITUATION 3 574.4 191.47 10.8207 5.960e-06 *** MONTH 1 1656.2 1656.20 93.6000 1.172e-14 *** SITUATION:MONTH 3 34.2 11.40 0.6443 0.5891 Residuals 72 1274.0 17.69 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(extract_log_lik(starling.rstan)))
Computed from 1500 by 80 log-likelihood matrix Estimate SE elpd_loo -233.7 5.0 p_loo 8.3 1.0 looic 467.4 10.0 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.# now fit a model without main factor modelString=" data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } ransformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,1000); sigma~cauchy(0,5); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " Xmat <- model.matrix(~SITUATION+MONTH, starling) starling.list <- with(starling,list(y=MASS, X=Xmat,n=nrow(starling), nX=ncol(Xmat))) starling.rstan.red <- stan(data=starling.list, model_code=modelString, chains=3, iter=2000, warmup=500, thin=3, refresh=FALSE )
Gradient evaluation took 4.8e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.48 seconds. Adjust your expectations accordingly! Elapsed Time: 0.071073 seconds (Warm-up) 0.106263 seconds (Sampling) 0.177336 seconds (Total) Gradient evaluation took 1.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds. Adjust your expectations accordingly! Elapsed Time: 0.069669 seconds (Warm-up) 0.100423 seconds (Sampling) 0.170092 seconds (Total) Gradient evaluation took 1.8e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.18 seconds. Adjust your expectations accordingly! Elapsed Time: 0.062487 seconds (Warm-up) 0.101526 seconds (Sampling) 0.164013 seconds (Total)(reduced=loo(extract_log_lik(starling.rstan.red)))
Computed from 1500 by 80 log-likelihood matrix Estimate SE elpd_loo -231.4 4.9 p_loo 5.5 0.7 looic 462.8 9.9 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
summary(starling.rstanarm)
Model Info: function: stan_glm family: gaussian [identity] formula: MASS ~ SITUATION * MONTH algorithm: sampling priors: see help('prior_summary') sample: 2250 (posterior sample size) num obs: 80 Estimates: mean sd 2.5% 25% 50% 75% 97.5% (Intercept) 90.8 1.3 88.2 89.9 90.8 91.7 93.4 SITUATIONS2 -0.6 1.9 -4.2 -1.9 -0.6 0.7 3.2 SITUATIONS3 -2.6 1.9 -6.3 -3.9 -2.6 -1.4 1.2 SITUATIONS4 -6.6 1.9 -10.3 -7.9 -6.7 -5.4 -2.9 MONTHNovember -7.3 1.9 -10.8 -8.5 -7.2 -6.1 -3.6 SITUATIONS2:MONTHNovember -3.6 2.7 -9.0 -5.3 -3.5 -1.8 1.8 SITUATIONS3:MONTHNovember -2.4 2.7 -7.8 -4.1 -2.3 -0.5 2.7 SITUATIONS4:MONTHNovember -1.6 2.7 -6.9 -3.4 -1.5 0.3 3.6 sigma 4.3 0.4 3.6 4.0 4.3 4.5 5.0 mean_PPD 83.8 0.7 82.5 83.3 83.8 84.3 85.1 log-posterior -245.3 2.3 -250.9 -246.5 -245.0 -243.6 -242.0 Diagnostics: mcse Rhat n_eff (Intercept) 0.0 1.0 1825 SITUATIONS2 0.0 1.0 1861 SITUATIONS3 0.0 1.0 2026 SITUATIONS4 0.0 1.0 1817 MONTHNovember 0.0 1.0 1809 SITUATIONS2:MONTHNovember 0.1 1.0 1915 SITUATIONS3:MONTHNovember 0.1 1.0 1800 SITUATIONS4:MONTHNovember 0.1 1.0 1745 sigma 0.0 1.0 1648 mean_PPD 0.0 1.0 1991 log-posterior 0.1 1.0 1416 For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).#OR library(broom) tidyMCMC(starling.rstanarm$stanfit,conf.int=TRUE, conf.method='HPDinterval', rhat=TRUE, ess=TRUE)
term estimate std.error conf.low conf.high rhat ess 1 (Intercept) 90.8342507 1.3482297 88.384550 93.596736 1.0004563 1825 2 SITUATIONS2 -0.6166141 1.9025010 -4.307238 3.045019 0.9997676 1861 3 SITUATIONS3 -2.6053800 1.9154558 -6.321085 1.223459 1.0017867 2026 4 SITUATIONS4 -6.6423833 1.9214525 -10.255593 -2.868400 0.9998412 1817 5 MONTHNovember -7.2506064 1.8911235 -10.800146 -3.553678 0.9997348 1809 6 SITUATIONS2:MONTHNovember -3.5640977 2.6910494 -8.963746 1.847141 0.9993705 1915 7 SITUATIONS3:MONTHNovember -2.3913123 2.7221593 -7.680779 2.833313 1.0015728 1800 8 SITUATIONS4:MONTHNovember -1.5670514 2.7163488 -6.709673 3.747383 0.9997508 1745 9 sigma 4.2750066 0.3606119 3.617221 5.001696 1.0004190 1648 10 mean_PPD 83.8022337 0.6747518 82.521057 85.120782 0.9992132 1991 11 log-posterior -245.2996925 2.2971964 -249.802970 -241.477210 1.0001864 1416
#OR with p-values newdata = with(starling, expand.grid(SITUATION=levels(SITUATION), MONTH=levels(MONTH))) Xmat = model.matrix(~SITUATION*MONTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(starling.rstanarm)[,i]) )
[1] 0 [1] 0.7488889 [1] 0.1657778 [1] 0.0004444444 [1] 0 [1] 0.1777778 [1] 0.3773333 [1] 0.5764444
# Main effect of SITUATION mcmcpvalue(as.matrix(starling.rstanarm)[,which(wch==1)])
[1] 0.004888889
# Main effect of Month mcmcpvalue(as.matrix(starling.rstanarm)[,which(wch==2)])
[1] 0
# Interaction mcmcpvalue(as.matrix(starling.rstanarm)[,which(wch==3)])
[1] 0.5968889
## frequentist for comparison summary(lm(MASS~SITUATION*MONTH, data=starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14anova(lm(MASS~SITUATION*MONTH, data=starling))
Analysis of Variance Table Response: MASS Df Sum Sq Mean Sq F value Pr(>F) SITUATION 3 574.4 191.47 10.8207 5.960e-06 *** MONTH 1 1656.2 1656.20 93.6000 1.172e-14 *** SITUATION:MONTH 3 34.2 11.40 0.6443 0.5891 Residuals 72 1274.0 17.69 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(starling.rstanarm))
Computed from 2250 by 80 log-likelihood matrix Estimate SE elpd_loo -233.6 5.0 p_loo 8.2 1.0 looic 467.1 10.0 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.starling.rstanarm.red = update(starling.rstanarm, .~SITUATION+MONTH)
Gradient evaluation took 4.6e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.46 seconds. Adjust your expectations accordingly! Elapsed Time: 0.117908 seconds (Warm-up) 0.377728 seconds (Sampling) 0.495636 seconds (Total) Gradient evaluation took 2.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.25 seconds. Adjust your expectations accordingly! Elapsed Time: 0.462271 seconds (Warm-up) 0.30388 seconds (Sampling) 0.766151 seconds (Total) Gradient evaluation took 2.2e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.22 seconds. Adjust your expectations accordingly! Elapsed Time: 0.179221 seconds (Warm-up) 0.276634 seconds (Sampling) 0.455855 seconds (Total)(reduced=loo(starling.rstanarm.red))
Computed from 2250 by 80 log-likelihood matrix Estimate SE elpd_loo -231.2 4.9 p_loo 5.3 0.6 looic 462.4 9.8 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
elpd_diff se 2.3 1.5There is very little support for an interaction. There are main effectssummary(starling.brms)
Family: gaussian(identity) Formula: MASS ~ SITUATION * MONTH Data: starling (Number of observations: 80) Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2; total post-warmup samples = 2250 ICs: LOO = NA; WAIC = NA; R2 = NA Population-Level Effects: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat Intercept 90.83 1.37 88.06 93.45 1533 1 SITUATIONS2 -0.64 1.97 -4.47 3.30 1573 1 SITUATIONS3 -2.64 1.91 -6.25 1.07 1455 1 SITUATIONS4 -6.62 1.95 -10.64 -2.81 1685 1 MONTHNovember -7.33 1.95 -11.25 -3.51 1503 1 SITUATIONS2:MONTHNovember -3.43 2.75 -8.92 1.89 1584 1 SITUATIONS3:MONTHNovember -2.21 2.68 -7.48 3.00 1579 1 SITUATIONS4:MONTHNovember -1.45 2.74 -6.79 4.08 1758 1 Family Specific Parameters: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat sigma 4.26 0.38 3.61 5.08 2026 1 Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat = 1).#OR library(broom) tidyMCMC(starling.brms$fit,conf.int=TRUE, conf.method='HPDinterval', rhat=TRUE, ess=TRUE)
term estimate std.error conf.low conf.high rhat ess 1 b_Intercept 90.8270337 1.3658808 88.241978 93.616386 1.0006760 1533 2 b_SITUATIONS2 -0.6438853 1.9659375 -4.899954 2.814581 1.0001759 1573 3 b_SITUATIONS3 -2.6449981 1.9064693 -6.267860 1.020327 1.0001993 1455 4 b_SITUATIONS4 -6.6181945 1.9474139 -10.445554 -2.692970 0.9998686 1685 5 b_MONTHNovember -7.3288521 1.9504259 -11.044855 -3.390492 1.0012561 1503 6 b_SITUATIONS2:MONTHNovember -3.4343794 2.7496251 -9.012966 1.772736 1.0007990 1584 7 b_SITUATIONS3:MONTHNovember -2.2091803 2.6833877 -7.424543 3.064406 1.0004977 1579 8 b_SITUATIONS4:MONTHNovember -1.4513879 2.7449835 -6.722984 4.129929 1.0001515 1758 9 sigma 4.2629104 0.3765409 3.522005 4.975503 1.0016642 2026
#OR with p-values newdata = with(starling, expand.grid(SITUATION=levels(SITUATION), MONTH=levels(MONTH))) Xmat = model.matrix(~SITUATION*MONTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(starling.brms)[,i]) )
[1] 0 [1] 0.736 [1] 0.1684444 [1] 0.001333333 [1] 0.0004444444 [1] 0.1942222 [1] 0.4071111 [1] 0.5848889
# Main effect of SITUATION mcmcpvalue(as.matrix(starling.brms)[,which(wch==1)])
[1] 0.004
# Main effect of Month mcmcpvalue(as.matrix(starling.brms)[,which(wch==2)])
[1] 0.0004444444
# Interaction mcmcpvalue(as.matrix(starling.brms)[,which(wch==3)])
[1] 0.6333333
## frequentist for comparison summary(lm(MASS~SITUATION*MONTH, data=starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14anova(lm(MASS~SITUATION*MONTH, data=starling))
Analysis of Variance Table Response: MASS Df Sum Sq Mean Sq F value Pr(>F) SITUATION 3 574.4 191.47 10.8207 5.960e-06 *** MONTH 1 1656.2 1656.20 93.6000 1.172e-14 *** SITUATION:MONTH 3 34.2 11.40 0.6443 0.5891 Residuals 72 1274.0 17.69 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(starling.brms))
LOOIC SE 467.43 10.11
starling.brms.red = update(starling.brms, .~SITUATION+MONTH, refresh=FALSE)
Gradient evaluation took 3.2e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.32 seconds. Adjust your expectations accordingly! Elapsed Time: 0.088189 seconds (Warm-up) 0.049832 seconds (Sampling) 0.138021 seconds (Total) Gradient evaluation took 1.2e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.12 seconds. Adjust your expectations accordingly! Elapsed Time: 0.098075 seconds (Warm-up) 0.060683 seconds (Sampling) 0.158758 seconds (Total) Gradient evaluation took 1.8e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.18 seconds. Adjust your expectations accordingly! Elapsed Time: 0.097611 seconds (Warm-up) 0.061486 seconds (Sampling) 0.159097 seconds (Total)(reduced=loo(starling.brms.red))
LOOIC SE 462.35 9.88
par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
- Explore the general effect of month across all situations
mcmc = starling.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^SITUATION|^MONTH", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 -9.096918 0.9460651 -10.88682 -7.148587
# OR if we express this as a percentage change Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 -10.2921 1.017045 -12.21395 -8.199951
mcmc = starling.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 -9.101269 0.961008 -11.01104 -7.228331
# OR if we express this as a percentage change Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 -10.29639 1.033416 -12.37168 -8.304203
mcmc = as.matrix(starling.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 -9.101405 0.9714053 -11.12969 -7.340376
# OR if we express this as a percentage change Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 -10.29556 1.044852 -12.4562 -8.387464
mcmc = as.matrix(starling.rstanarm) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SITUATION|^MONTH',colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 -9.131222 0.955334 -11.08878 -7.362893
# OR if we express this as a percentage change Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 -10.328 1.027833 -12.42573 -8.414559
mcmc = as.matrix(starling.brms) wch = grep('^b_',colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 -9.102589 0.9715485 -11.14465 -7.296226
# OR if we express this as a percentage change Xmat = model.matrix(~SITUATION*MONTH,newdata) Xmat = aggregate(Xmat, list(newdata$MONTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 -10.29743 1.044987 -12.3539 -8.228035
- Generate a summary figure
mcmc = starling.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^SITUATION|^MONTH", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
SITUATION MONTH estimate std.error conf.low conf.high 1 S1 January 90.79215 1.356498 88.21102 93.49823 2 S2 January 90.18891 1.366450 87.54662 92.89458 3 S3 January 88.20137 1.344198 85.46808 90.76530 4 S4 January 84.19057 1.348469 81.59279 86.88510 5 S1 November 83.58889 1.338832 81.00680 86.21545 6 S2 November 79.38717 1.354513 76.72113 82.01367 7 S3 November 78.59354 1.368455 75.90668 81.25595 8 S4 November 75.41573 1.347574 72.84158 78.10217
ggplot(newdata, aes(y=estimate, x=SITUATION, fill=MONTH)) + geom_blank() + geom_line(aes(x=as.numeric(SITUATION), linetype=MONTH)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=MONTH), size=3)+ scale_y_continuous('Mass (g)')+ scale_x_discrete('Situation')+ scale_shape_manual('Month',values=c(21,16))+ scale_fill_manual('Month',values=c('white','black'))+ scale_linetype_manual('Month',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

mcmc = starling.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
SITUATION MONTH estimate std.error conf.low conf.high 1 S1 January 90.78873 1.358058 88.09834 93.39648 2 S2 January 90.19961 1.358988 87.48714 92.84222 3 S3 January 88.19100 1.340393 85.58381 90.90540 4 S4 January 84.21058 1.360060 81.47964 86.87083 5 S1 November 83.59256 1.353514 80.91467 86.26002 6 S2 November 79.40225 1.345935 76.72388 81.99603 7 S3 November 78.59046 1.364822 75.86100 81.23437 8 S4 November 75.39957 1.362908 72.76250 78.10410
ggplot(newdata, aes(y=estimate, x=SITUATION, fill=MONTH)) + geom_blank() + geom_line(aes(x=as.numeric(SITUATION), linetype=MONTH)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=MONTH), size=3)+ scale_y_continuous('Mass (g)')+ scale_x_discrete('Situation')+ scale_shape_manual('Month',values=c(21,16))+ scale_fill_manual('Month',values=c('white','black'))+ scale_linetype_manual('Month',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

mcmc = as.matrix(starling.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) Xmat = model.matrix(~SITUATION*MONTH,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
SITUATION MONTH estimate std.error conf.low conf.high 1 S1 January 90.80079 1.350603 88.16097 93.33563 2 S2 January 90.19866 1.366092 87.53747 92.77494 3 S3 January 88.23510 1.409559 85.60655 90.99940 4 S4 January 84.18831 1.408740 81.40267 86.81393 5 S1 November 83.58597 1.345420 81.07045 86.35281 6 S2 November 79.35734 1.357414 76.91119 82.14984 7 S3 November 78.60834 1.375153 76.04264 81.39771 8 S4 November 75.46560 1.354976 72.79973 78.20676
ggplot(newdata, aes(y=estimate, x=SITUATION, fill=MONTH)) + geom_blank() + geom_line(aes(x=as.numeric(SITUATION), linetype=MONTH)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=MONTH), size=3)+ scale_y_continuous('Mass (g)')+ scale_x_discrete('Situation')+ scale_shape_manual('Month',values=c(21,16))+ scale_fill_manual('Month',values=c('white','black'))+ scale_linetype_manual('Month',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

newdata = expand.grid(SITUATION=levels(starling$SITUATION), MONTH=levels(starling$MONTH)) fit = posterior_linpred(starling.rstanarm, newdata = newdata) newdata = newdata %>% cbind(tidyMCMC(as.mcmc(fit),conf.int = TRUE,conf.method = "HPDinterval")) ggplot(newdata, aes(y=estimate, x=SITUATION, fill=MONTH)) + geom_blank() + geom_line(aes(x=as.numeric(SITUATION), linetype=MONTH)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high))+ geom_point(aes(shape=MONTH), size=3)+ scale_y_continuous('Mass (g)')+ scale_x_discrete('Situation')+ scale_shape_manual('Month',values=c(21,16))+ scale_fill_manual('Month',values=c('white','black'))+ scale_linetype_manual('Month',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

## The simple way plot(marginal_effects(starling.brms))



## OR eff=marginal_effects(starling.brms) ggplot(eff[['SITUATION:MONTH']], aes(y=estimate__, x=SITUATION, fill=MONTH)) + geom_blank() + geom_line(aes(x=as.numeric(SITUATION), linetype=MONTH)) + geom_linerange(aes(ymin=lower__, ymax=upper__))+ geom_point(aes(shape=MONTH), size=3)+ scale_y_continuous('Y')+ scale_x_discrete('Situation')+ scale_shape_manual('Month',values=c(21,16))+ scale_fill_manual('Month',values=c('white','black'))+ scale_linetype_manual('Month',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

- Explore finite-population standard deviations
mcmc = starling.mcmcpack Xmat = model.matrix(~SITUATION*MONTH, starling) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SITUATION = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.MONTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SITUATION|^MONTH',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,starling$MASS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SITUATION, sd.MONTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 3.188110 0.7490819 1.6903288 4.625065 2 sd.MONTH 5.093515 1.3383360 2.4484730 7.728094 3 sd.Int 2.105215 0.9017623 0.4779652 3.875961 4 sd.resid 4.211216 0.1077824 4.0383643 4.417717
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 21.92458 3.810305 13.815261 28.70201 2 sd.MONTH 35.37060 7.395632 20.109079 48.01344 3 sd.Int 13.68865 6.512478 2.970593 27.39771 4 sd.resid 29.04577 2.705921 23.732845 34.40443
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = starling.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~SITUATION*MONTH, starling) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SITUATION = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.MONTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,starling$MASS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SITUATION, sd.MONTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 3.179145 0.7526149 1.6901694 4.642912 2 sd.MONTH 5.088492 1.3555137 2.5101245 7.809579 3 sd.Int 2.112234 0.9068484 0.4613288 3.861946 4 sd.resid 4.211964 0.1087270 4.0417787 4.425289
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 21.87844 3.824123 13.802679 28.59839 2 sd.MONTH 35.46236 7.492723 19.187885 47.29183 3 sd.Int 13.71059 6.590495 3.009805 27.33991 4 sd.resid 29.06871 2.735405 23.654424 34.56475
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(starling.rstan) Xmat = model.matrix(~SITUATION*MONTH, starling) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SITUATION = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.MONTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,starling$MASS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SITUATION, sd.MONTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 3.204405 0.7589083 1.7690046 4.725067 2 sd.MONTH 5.101650 1.3417947 2.5345140 7.801372 3 sd.Int 2.130984 0.9018745 0.3659416 3.834433 4 sd.resid 4.215527 0.1141759 4.0374031 4.429020
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 22.01324 3.822899 14.307682 29.26592 2 sd.MONTH 35.45012 7.385197 18.576168 46.67093 3 sd.Int 13.85862 6.479191 2.620738 27.59213 4 sd.resid 28.84171 2.755158 23.935655 34.60721
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(starling.rstanarm) Xmat = model.matrix(~SITUATION*MONTH, starling) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SITUATION = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.MONTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SITUATION|^MONTH',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,starling$MASS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SITUATION, sd.MONTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 3.201467 0.7552157 1.634315 4.590631 2 sd.MONTH 5.126953 1.3372263 2.512830 7.636856 3 sd.Int 2.093080 0.9183301 0.512600 4.008883 4 sd.resid 4.213128 0.1106179 4.046633 4.442038
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 21.94638 3.809934 14.112844 28.97649 2 sd.MONTH 35.47365 7.390306 20.475241 48.42700 3 sd.Int 13.45762 6.626827 1.942844 27.06199 4 sd.resid 28.93120 2.757289 23.348478 34.21859
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(starling.brms) Xmat = model.matrix(~SITUATION*MONTH, starling) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SITUATION = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.MONTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^b_',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,starling$MASS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SITUATION, sd.MONTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 3.190988 0.7523201 1.7128319 4.651159 2 sd.MONTH 5.182281 1.3791594 2.3974400 7.809892 3 sd.Int 2.050548 0.9010897 0.4109023 3.766794 4 sd.resid 4.212525 0.1116127 4.0366221 4.429444
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SITUATION 21.93673 3.826436 14.029390 28.58547 2 sd.MONTH 35.75830 7.519683 19.507211 48.07652 3 sd.Int 13.17520 6.492255 2.673058 26.83970 4 sd.resid 29.03860 2.707223 23.628654 34.34458
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

- Estimate a psuedo-$R^2$
library(broom) mcmc <- starling.mcmcpack Xmat = model.matrix(~SITUATION*MONTH, starling) wch = c(which(colnames(mcmc)=='(Intercept)'), grep('^SITUATION|^MONTH', colnames(mcmc))) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, starling$MASS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.6262851 0.04079205 0.5439231 0.6967781
#for comparison with frequentist summary(lm(MASS ~ SITUATION*MONTH, starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14library(broom) mcmc <- starling.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~SITUATION*MONTH, starling) wch = grep('^beta', colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, starling$MASS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.6262546 0.04166799 0.5434732 0.6984009
#for comparison with frequentist summary(lm(MASS ~ SITUATION*MONTH, starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14library(broom) mcmc <- as.matrix(starling.rstan) Xmat = model.matrix(~SITUATION*MONTH, starling) wch = grep('^beta', colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, starling$MASS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.6256618 0.04176619 0.5400687 0.6952968
#for comparison with frequentist summary(lm(MASS ~ SITUATION*MONTH, starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14library(broom) mcmc <- as.matrix(starling.rstanarm) Xmat = model.matrix(~SITUATION*MONTH, starling) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SITUATION|^MONTH',colnames(mcmc))) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, starling$MASS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.6277337 0.04180136 0.5469394 0.6982494
#for comparison with frequentist summary(lm(MASS ~ SITUATION*MONTH, starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14library(broom) mcmc <- as.matrix(starling.brms) Xmat = model.matrix(~SITUATION*MONTH, starling) wch = grep('^b_',colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, starling$MASS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.6253397 0.04208681 0.5440957 0.7028489
#for comparison with frequentist summary(lm(MASS ~ SITUATION*MONTH, starling))
Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.800 1.330 68.260 < 2e-16 *** SITUATIONS2 -0.600 1.881 -0.319 0.750691 SITUATIONS3 -2.600 1.881 -1.382 0.171213 SITUATIONS4 -6.600 1.881 -3.508 0.000781 *** MONTHNovember -7.200 1.881 -3.827 0.000274 *** SITUATIONS2:MONTHNovember -3.600 2.660 -1.353 0.180233 SITUATIONS3:MONTHNovember -2.400 2.660 -0.902 0.370003 SITUATIONS4:MONTHNovember -1.600 2.660 -0.601 0.549455 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.206 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.28 on 7 and 72 DF, p-value: 9.546e-14
Unbalanced Two-factor ANOVA
Here is a modified example from Quinn and Keough (2002). Stehman and Meredith (1995) present data from an experiment that was set up to test the hypothesis that healthy spruce seedlings break bud sooner than diseased spruce seedlings. There were 2 factors: pH (3 levels: 3, 5.5, 7) and HEALTH (2 levels: healthy, diseased). The dependent variable was the average (from 5 buds) bud emergence rating (BRATING) on each seedling. The sample size varied for each combination of pH and health, ranging from 7 to 23 seedlings. With two factors, this experiment should be analyzed with a 2 factor (2 x 3) ANOVA.
Download Stehman data set| Format of stehman.csv data files | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
stehman <- read.table("../downloads/data/stehman.csv", header = T, sep = ",", strip.white = T) head(stehman)
PH HEALTH GROUP BRATING 1 3 D D3 0.0 2 3 D D3 0.8 3 3 D D3 0.8 4 3 D D3 0.8 5 3 D D3 0.8 6 3 D D3 0.8
The variable PH contains a list of pH values and is supposed to represent a factorial variable. However, because the contents of this variable are numbers, R initially treats them as numbers, and therefore considers the variable to be numeric rather than categorical. In order to force R/JAGS to treat this variable as a factor (categorical) it must be in categorical form (yet as numbers). Confused? What this means, is that to be treated as a factor, its levels must be indices and therefore the PH variable needs to be converted into a factor before being converted to a numeric. That way the levels 3, 5.5 and 7 will be coded as 1, 2 and 3.
stehman = stehman %>% mutate(PH = factor(PH))
Exploratory data analysis did not reveal any issues with normality or homogeneity of variance.
- Fit the model to investigate the effects of pH and health status on
the bud emergence rating of spruce seedlings.
$$
\begin{align}
y_{ij} &\sim{} N(\mu_{ij}, \sigma)\\
\mu_{ij} &= \beta_0 + \mathbf{X}\boldsymbol{\beta}\\[1em]
\beta_0 &\sim{} N(0,10)\\
\beta &\sim{} N(0,1)\\
\sigma &\sim{} cauchy(0,2)\\
\end{align}
$$
library(MCMCpack) stehman.mcmcpack = MCMCregress(BRATING ~ PH * HEALTH, data = stehman)
modelString = " model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(beta[],X[i,]) } #Priors beta0 ~ dnorm(0.01,1.0E-6) for (j in 1:nX) { beta[j] ~ dnorm(0.01,1.0E-6) } tau <- 1 / (sigma * sigma) sigma~dunif(0,100) } " X = model.matrix(~PH * HEALTH, data = stehman) stehman.list <- with(stehman, list(y = BRATING, X = X, nX = ncol(X), n = nrow(stehman))) params <- c("beta", "sigma") burnInSteps = 3000 nChains = 3 numSavedSteps = 15000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) stehman.r2jags <- jags(data = stehman.list, inits = NULL, parameters.to.save = params, model.file = textConnection(modelString), n.chains = nChains, n.iter = nIter, n.burnin = burnInSteps, n.thin = thinSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph information: Observed stochastic nodes: 95 Unobserved stochastic nodes: 8 Total graph size: 796 Initializing model
modelString = " data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } transformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,10); sigma~cauchy(0,2); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " X = model.matrix(~PH * HEALTH, data = stehman) stehman.list <- with(stehman, list(y = BRATING, X = X, nX = ncol(X), n = nrow(stehman))) stehman.rstan <- stan(data = stehman.list, model_code = modelString, chains = 3, iter = 2000, warmup = 500, thin = 3)
In file included from /usr/local/lib/R/site-library/BH/include/boost/config.hpp:39:0, from /usr/local/lib/R/site-library/BH/include/boost/math/tools/config.hpp:13, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/var.hpp:7, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/gevv_vvv_vari.hpp:5, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core.hpp:12, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/mat.hpp:4, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math.hpp:4, from /usr/local/lib/R/site-library/StanHeaders/include/src/stan/model/model_header.hpp:4, from file71d5471c7e.cpp:8: /usr/local/lib/R/site-library/BH/include/boost/config/compiler/gcc.hpp:186:0: warning: "BOOST_NO_CXX11_RVALUE_REFERENCES" redefined # define BOOST_NO_CXX11_RVALUE_REFERENCES ^ <command-line>:0:0: note: this is the location of the previous definition SAMPLING FOR MODEL 'fa2805f19ebd689dd8d4714ae0df073d' NOW (CHAIN 1). Gradient evaluation took 5.2e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.52 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.044153 seconds (Warm-up) 0.120243 seconds (Sampling) 0.164396 seconds (Total) SAMPLING FOR MODEL 'fa2805f19ebd689dd8d4714ae0df073d' NOW (CHAIN 2). Gradient evaluation took 1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.045313 seconds (Warm-up) 0.124027 seconds (Sampling) 0.16934 seconds (Total) SAMPLING FOR MODEL 'fa2805f19ebd689dd8d4714ae0df073d' NOW (CHAIN 3). Gradient evaluation took 1.1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.046914 seconds (Warm-up) 0.120826 seconds (Sampling) 0.16774 seconds (Total)print(stehman.rstan, par = c("beta", "sigma"))
Inference for Stan model: fa2805f19ebd689dd8d4714ae0df073d. 3 chains, each with iter=2000; warmup=500; thin=3; post-warmup draws per chain=500, total post-warmup draws=1500. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.19 0.00 0.11 0.97 1.12 1.19 1.26 1.40 1304 1 beta[2] -0.38 0.00 0.15 -0.67 -0.49 -0.39 -0.28 -0.09 1374 1 beta[3] -0.07 0.00 0.16 -0.36 -0.18 -0.07 0.04 0.24 1330 1 beta[4] 0.42 0.01 0.19 0.04 0.30 0.42 0.54 0.78 1293 1 beta[5] 0.00 0.01 0.30 -0.59 -0.20 -0.01 0.20 0.58 1436 1 beta[6] -0.20 0.01 0.27 -0.72 -0.39 -0.20 -0.02 0.33 1204 1 sigma 0.51 0.00 0.04 0.44 0.49 0.51 0.54 0.60 1282 1 Samples were drawn using NUTS(diag_e) at Mon Dec 18 12:47:51 2017. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).stehman.rstanarm = stan_glm(BRATING ~ PH * HEALTH, data = stehman, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior_intercept = normal(0, 10), prior = normal(0, 1), prior_aux = cauchy(0, 2))
Gradient evaluation took 4.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.44 seconds. Adjust your expectations accordingly! Elapsed Time: 0.07264 seconds (Warm-up) 0.158764 seconds (Sampling) 0.231404 seconds (Total) Gradient evaluation took 1.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds. Adjust your expectations accordingly! Elapsed Time: 0.068386 seconds (Warm-up) 0.155984 seconds (Sampling) 0.22437 seconds (Total) Gradient evaluation took 1.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds. Adjust your expectations accordingly! Elapsed Time: 0.075019 seconds (Warm-up) 0.157612 seconds (Sampling) 0.232631 seconds (Total)print(stehman.rstanarm)
stan_glm family: gaussian [identity] formula: BRATING ~ PH * HEALTH ------ Estimates: Median MAD_SD (Intercept) 1.2 0.1 PH5.5 -0.4 0.1 PH7 -0.1 0.1 HEALTHH 0.4 0.2 PH5.5:HEALTHH 0.0 0.2 PH7:HEALTHH -0.1 0.2 sigma 0.5 0.0 Sample avg. posterior predictive distribution of y (X = xbar): Median MAD_SD mean_PPD 1.2 0.1 ------ For info on the priors used see help('prior_summary.stanreg').tidyMCMC(stehman.rstanarm, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 (Intercept) 1.19367197 0.09811005 1.00267082 1.3794144 2 PH5.5 -0.37103685 0.13716153 -0.62890963 -0.1015450 3 PH7 -0.07738565 0.14394092 -0.36568565 0.2005097 4 HEALTHH 0.38310472 0.16157074 0.05041117 0.6909403 5 PH5.5:HEALTHH 0.01888488 0.24248171 -0.46984092 0.4773634 6 PH7:HEALTHH -0.14853772 0.23370558 -0.58939364 0.3265783 7 sigma 0.51176627 0.03809169 0.44268636 0.5919980
stehman.brms = brm(BRATING ~ PH * HEALTH, data = stehman, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior = c(prior(normal(0, 10), class = "Intercept"), prior(normal(0, 1), class = "b"), prior(cauchy(0, 2), class = "sigma")))
Gradient evaluation took 2.9e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.29 seconds. Adjust your expectations accordingly! Elapsed Time: 0.039309 seconds (Warm-up) 0.110191 seconds (Sampling) 0.1495 seconds (Total) Gradient evaluation took 1.1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds. Adjust your expectations accordingly! Elapsed Time: 0.041103 seconds (Warm-up) 0.098148 seconds (Sampling) 0.139251 seconds (Total) Gradient evaluation took 1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds. Adjust your expectations accordingly! Elapsed Time: 0.039301 seconds (Warm-up) 0.108025 seconds (Sampling) 0.147326 seconds (Total)print(stehman.brms)
Family: gaussian(identity) Formula: BRATING ~ PH * HEALTH Data: stehman (Number of observations: 95) Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2; total post-warmup samples = 2250 ICs: LOO = NA; WAIC = NA; R2 = NA Population-Level Effects: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat Intercept 1.19 0.11 0.98 1.40 1807 1 PH5.5 -0.38 0.15 -0.67 -0.08 1731 1 PH7 -0.07 0.15 -0.38 0.22 1911 1 HEALTHH 0.42 0.18 0.06 0.78 1797 1 PH5.5:HEALTHH 0.01 0.28 -0.53 0.55 1392 1 PH7:HEALTHH -0.19 0.26 -0.69 0.33 1767 1 Family Specific Parameters: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat sigma 0.51 0.04 0.45 0.6 2144 1 Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat = 1).tidyMCMC(stehman.brms, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 b_Intercept 1.193674756 0.10530085 0.99311929 1.40494393 2 b_PH5.5 -0.381747258 0.14726079 -0.68375335 -0.09339214 3 b_PH7 -0.071524112 0.15253709 -0.38083958 0.21668439 4 b_HEALTHH 0.417002902 0.18254053 0.06053687 0.78468311 5 b_PH5.5:HEALTHH 0.007985665 0.27642096 -0.52552795 0.55176171 6 b_PH7:HEALTHH -0.194003633 0.26341846 -0.69066979 0.32520555 7 sigma 0.512449370 0.03793437 0.44254238 0.59053416
- Explore MCMC diagnostics
library(MCMCpack) plot(stehman.mcmcpack)


raftery.diag(stehman.mcmcpack)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) (Intercept) 2 3834 3746 1.020 PH5.5 2 3620 3746 0.966 PH7 2 3710 3746 0.990 HEALTHH 2 3710 3746 0.990 PH5.5:HEALTHH 2 3802 3746 1.010 PH7:HEALTHH 2 3771 3746 1.010 sigma2 2 3931 3746 1.050autocorr.diag(stehman.mcmcpack)
(Intercept) PH5.5 PH7 HEALTHH PH5.5:HEALTHH PH7:HEALTHH Lag 0 1.0000000000 1.000000000 1.000000000 1.0000000000 1.000000000 1.000000000 Lag 1 -0.0082664884 -0.004293283 -0.012050219 -0.0002933288 -0.011085366 0.005274514 Lag 5 0.0211074297 0.021791290 -0.006542664 0.0067078263 0.014878737 -0.013265007 Lag 10 0.0118041625 0.019271612 -0.003964872 -0.0097575506 0.008702777 -0.007317416 Lag 50 -0.0008092011 -0.008724041 0.016744280 -0.0040749327 0.003223703 0.003821559 sigma2 Lag 0 1.0000000000 Lag 1 0.0739011931 Lag 5 0.0060177813 Lag 10 0.0163620083 Lag 50 0.0007361694stehman.mcmc = as.mcmc(stehman.r2jags) plot(stehman.mcmc)


preds <- grep("beta", colnames(stehman.mcmc[[1]])) plot(stehman.mcmc[, preds])


raftery.diag(stehman.mcmc)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 37020 3746 9.88 beta[2] 20 38330 3746 10.20 beta[3] 20 38330 3746 10.20 beta[4] 20 38330 3746 10.20 beta[5] 20 37020 3746 9.88 beta[6] 20 37020 3746 9.88 deviance 30 40390 3746 10.80 sigma 20 38330 3746 10.20 [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 38330 3746 10.20 beta[2] 20 37020 3746 9.88 beta[3] 10 37660 3746 10.10 beta[4] 20 39000 3746 10.40 beta[5] 20 36380 3746 9.71 beta[6] 20 39680 3746 10.60 deviance 20 37020 3746 9.88 sigma 20 36380 3746 9.71 [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 38330 3746 10.20 beta[2] 20 37020 3746 9.88 beta[3] 20 37020 3746 9.88 beta[4] 20 37020 3746 9.88 beta[5] 20 39680 3746 10.60 beta[6] 20 38330 3746 10.20 deviance 10 37660 3746 10.10 sigma 20 37020 3746 9.88autocorr.diag(stehman.mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] deviance Lag 0 1.000000000 1.000000000 1.000000000 1.000000000 1.000000e+00 1.000000000 1.000000000 Lag 10 -0.000476526 0.005629233 -0.001526660 0.003940911 -1.394040e-02 0.001486377 -0.001298144 Lag 50 0.003017091 -0.002427786 0.003100058 -0.003805200 4.121381e-03 -0.009445500 -0.014443303 Lag 100 0.014172346 0.013514742 0.002521652 -0.011685309 -2.373411e-05 -0.014611819 -0.026508639 Lag 500 -0.003974150 0.004348333 0.010319143 0.011644278 3.585185e-04 0.003634270 0.001901838 sigma Lag 0 1.000000000 Lag 10 -0.011846387 Lag 50 0.007921908 Lag 100 0.013128371 Lag 500 -0.005813782s = as.array(stehman.rstan) wch = grep("beta", dimnames(s)$parameters) s = s[, , wch] mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)


autocorr.diag(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] Lag 0 1.00000000 1.000000000 1.0000000000 1.000000e+00 1.00000000 Lag 1 0.06897619 0.042973255 0.0558674722 7.510733e-02 0.02016133 Lag 5 -0.01934273 0.008356967 -0.0010678286 1.597363e-02 -0.00913322 Lag 10 0.01386228 0.027487992 0.0004694462 -1.746176e-03 0.03028257 Lag 50 -0.01452068 0.021524605 -0.0058331040 8.032396e-05 0.03676614
## Or via rstan stan_trace(stehman.rstan)

stan_ac(stehman.rstan)

stan_rhat(stehman.rstan)

stan_ess(stehman.rstan)

## Or via bayesplot detach("package:reshape") mcmc_trace(as.matrix(stehman.rstan), regex_pars = "beta|sigma")

mcmc_dens(as.matrix(stehman.rstan), regex_pars = "beta|sigma")

s = as.array(stehman.rstanarm) mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)


autocorr.diag(mcmc)
(Intercept) PH5.5 PH7 HEALTHH PH5.5:HEALTHH PH7:HEALTHH Lag 0 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 Lag 1 0.072672217 0.035955905 0.041920277 0.098061797 0.089104134 0.089477153 Lag 5 0.001942657 -0.042864644 0.001999419 -0.003831644 -0.009633957 -0.003776616 Lag 10 0.023266856 0.022048618 -0.004042822 0.007077818 0.027393777 0.012352532 Lag 50 0.004084505 0.005365237 0.019703330 0.022985703 0.001591890 0.002965411
## OR via rstan stan_trace(stehman.rstanarm)

raftery.diag(stehman.rstanarm)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
stan_ac(stehman.rstanarm)

stan_rhat(stehman.rstanarm)

stan_ess(stehman.rstanarm)

## OR via bayesplot detach("package:reshape") mcmc_trace(as.array(stehman.rstanarm), regex_pars = "Intercept|PH|HEALTH|sigma")

mcmc_dens(as.array(stehman.rstanarm))

posterior_vs_prior(stehman.rstanarm, color_by = "vs", group_by = TRUE, facet_args = list(scales = "free_y"))
Gradient evaluation took 2.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.24 seconds. Adjust your expectations accordingly! Elapsed Time: 0.043561 seconds (Warm-up) 0.126228 seconds (Sampling) 0.169789 seconds (Total) Gradient evaluation took 1.7e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds. Adjust your expectations accordingly! Elapsed Time: 0.047312 seconds (Warm-up) 0.100026 seconds (Sampling) 0.147338 seconds (Total)
mcmc = as.mcmc(stehman.brms) plot(mcmc)


autocorr.diag(mcmc)
Error in ts(x, start = start(x), end = end(x), deltat = thin(x)): invalid time series parameters specified
## OR via rstan stan_trace(stehman.brms$fit)

raftery.diag(stehman.brms)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
stan_ac(stehman.brms$fit)

stan_rhat(stehman.brms$fit)

stan_ess(stehman.brms$fit)

- Explore model validation
mcmc = as.data.frame(stehman.mcmcpack) #generate a model matrix newdata = stehman Xmat = model.matrix(~PH*HEALTH, newdata) ##get median parameter estimates head(mcmc)
(Intercept) PH5.5 PH7 HEALTHH PH5.5:HEALTHH PH7:HEALTHH sigma2 1 1.1958607 -0.2466107 -0.01938051 0.3556211 -0.2893223 0.02019784 0.2876577 2 1.1910616 -0.2431350 -0.04375726 0.6126346 -0.2114437 -0.38413965 0.2569897 3 1.1488226 -0.3306775 0.10781837 0.6846604 -0.1645004 -0.48241555 0.2160724 4 0.8847077 -0.2066633 0.19193643 0.5555766 0.1136555 -0.30156261 0.2617900 5 1.1890155 -0.2925227 -0.02105352 0.4561701 0.0288746 -0.34923711 0.2592518 6 1.0998312 -0.3455959 -0.02276005 0.6807667 -0.2297119 -0.43344812 0.2297707
wch = grepl('sigma2',colnames(mcmc))==0 coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = stehman$BRATING - fit ggplot() + geom_point(stehman=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=PH))

ggplot(newdata) + geom_point(aes(y=resid, x=HEALTH))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model wch = grepl('sigma',colnames(mcmc))==0 coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~PH*HEALTH, data=stehman) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(stehman), fit[i,], sqrt(mcmc[i, 'sigma2']))) newdata = data.frame(PH=stehman$PH, HEALTH=stehman$HEALTH, yRep) %>% gather(key=Sample, value=Value,-PH,-HEALTH) ggplot(newdata) + geom_violin(aes(y=Value, x=PH, color=HEALTH,fill='Model'), alpha=0.5)+ geom_violin(data=stehman, aes(y=BRATING,x=PH,color=HEALTH,fill='Obs'), alpha=0.5) + geom_point(data=stehman, aes(y=BRATING, x=PH, color=HEALTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=HEALTH, fill='Model', group=HEALTH, color=PH), alpha=0.5)+ geom_point(data=stehman, aes(y=BRATING, x=HEALTH, group=HEALTH,color=PH))

mcmc_intervals(as.matrix(stehman.mcmcpack), regex_pars='Intercept|^PH|^HEALTH|sigma')

mcmc_areas(as.matrix(stehman.mcmcpack), regex_pars='Intercept|^PH|^HEALTH|sigma')

mcmc = stehman.r2jags$BUGSoutput$sims.matrix #generate a model matrix newdata = stehman Xmat = model.matrix(~PH*HEALTH, newdata) ##get median parameter estimates head(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] deviance sigma [1,] 1.081489 -0.3547513 0.12080492 0.8130361 -0.24347937 -0.5478541 142.3056 0.4621175 [2,] 1.275060 -0.2830531 -0.08333704 0.5522064 -0.58752129 -0.4894594 143.6771 0.4746738 [3,] 1.186234 -0.5703387 -0.19082916 0.4528418 0.03605714 -0.2869267 140.8598 0.5221374 [4,] 1.178655 -0.3164378 0.01123911 0.5127060 -0.27971191 -0.5073551 136.5782 0.5106601 [5,] 1.381500 -0.7378375 -0.17605800 0.4327969 0.01448500 -0.3437259 145.0874 0.4613809 [6,] 1.211761 -0.4856985 -0.20125901 0.4415108 -0.28223115 -0.1664190 144.5723 0.5999335
wch = grep('^beta\\[',colnames(mcmc)) coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = stehman$BRATING - fit ggplot() + geom_point(stehman=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=PH))

ggplot(newdata) + geom_point(aes(y=resid, x=HEALTH))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~PH*HEALTH, data=stehman) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(stehman), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(PH=stehman$PH, HEALTH=stehman$HEALTH, yRep) %>% gather(key=Sample, value=Value,-PH,-HEALTH) ggplot(newdata) + geom_violin(aes(y=Value, x=PH, color=HEALTH,fill='Model'), alpha=0.5)+ geom_violin(data=stehman, aes(y=BRATING,x=PH,color=HEALTH,fill='Obs'), alpha=0.5) + geom_point(data=stehman, aes(y=BRATING, x=PH, color=HEALTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=HEALTH, fill='Model', group=HEALTH, color=PH), alpha=0.5)+ geom_point(data=stehman, aes(y=BRATING, x=HEALTH, group=HEALTH,color=PH))

mcmc_intervals(as.matrix(stehman.mcmcpack), regex_pars='Intercept|^PH|^HEALTH|sigma')

mcmc_areas(as.matrix(stehman.mcmcpack), regex_pars='Intercept|^PH|^HEALTH|sigma')

mcmc = as.matrix(stehman.rstan) #generate a model matrix newdata = stehman Xmat = model.matrix(~PH*HEALTH, newdata) ##get median parameter estimates wch = grep('^beta\\[',colnames(mcmc)) coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = stehman$BRATING - fit ggplot() + geom_point(stehman=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=PH))

ggplot(newdata) + geom_point(aes(y=resid, x=HEALTH))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~PH*HEALTH, data=stehman) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(stehman), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(PH=stehman$PH, HEALTH=stehman$HEALTH, yRep) %>% gather(key=Sample, value=Value,-PH,-HEALTH) ggplot(newdata) + geom_violin(aes(y=Value, x=PH, color=HEALTH,fill='Model'), alpha=0.5)+ geom_violin(data=stehman, aes(y=BRATING,x=PH,color=HEALTH,fill='Obs'), alpha=0.5) + geom_point(data=stehman, aes(y=BRATING, x=PH, color=HEALTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=HEALTH, fill='Model', group=HEALTH, color=PH), alpha=0.5)+ geom_point(data=stehman, aes(y=BRATING, x=HEALTH, group=HEALTH,color=PH))

mcmc_intervals(as.matrix(stehman.rstan), regex_pars='^beta|sigma')

mcmc_areas(as.matrix(stehman.rstan), regex_pars='^beta|sigma')

resid = resid(stehman.rstanarm) fit = fitted(stehman.rstanarm) ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

resid = resid(stehman.rstanarm) dat = stehman %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = PH))

ggplot(dat) + geom_point(aes(y = resid, x = HEALTH))

resid = resid(stehman.rstanarm) sigma(stehman.rstanarm)
[1] 0.5094637
sresid = resid/sigma(stehman.rstanarm) fit = fitted(stehman.rstanarm) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

y_pred = posterior_predict(stehman.rstanarm) newdata = stehman %>% dplyr:::select(-GROUP) %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -PH,-HEALTH,-BRATING) head(newdata)
PH HEALTH BRATING Rep Value 1 3 D 0.0 1 0.8358639 2 3 D 0.8 1 1.1077990 3 3 D 0.8 1 1.0122379 4 3 D 0.8 1 1.6161427 5 3 D 0.8 1 1.9839090 6 3 D 0.8 1 2.1736836
ggplot(newdata) + geom_violin(aes(y=Value, x=PH, color=HEALTH,fill='Model'), alpha=0.5)+ geom_violin(data=stehman, aes(y=BRATING,x=PH,color=HEALTH,fill='Obs'), alpha=0.5) + geom_point(data=stehman, aes(y=BRATING, x=PH, color=HEALTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=HEALTH, fill='Model', group=HEALTH, color=PH), alpha=0.5)+ geom_point(data=stehman, aes(y=BRATING, x=HEALTH, group=HEALTH,color=PH))

mcmc_intervals(as.matrix(stehman.rstanarm), regex_pars='Intercept|^PH|^HEALTH|sigma')

mcmc_areas(as.matrix(stehman.rstanarm), regex_pars='Intercept|^PH|^HEALTH|sigma')
 All diagnostics seem reasonable.
All diagnostics seem reasonable.resid = resid(stehman.brms)[,'Estimate'] fit = fitted(stehman.brms)[,'Estimate'] ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

resid = resid(stehman.brms)[,'Estimate'] dat = stehman %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = PH))

ggplot(dat) + geom_point(aes(y = resid, x = HEALTH))

resid = resid(stehman.brms) sresid = resid(stehman.brms, type='pearson')[,'Estimate'] fit = fitted(stehman.brms)[,'Estimate'] ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

y_pred = posterior_predict(stehman.brms) newdata = stehman %>% dplyr:::select(-GROUP) %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -PH,-HEALTH,-BRATING) head(newdata)
PH HEALTH BRATING Rep Value 1 3 D 0.0 1 1.2650494 2 3 D 0.8 1 0.5353851 3 3 D 0.8 1 1.6889294 4 3 D 0.8 1 1.6245131 5 3 D 0.8 1 0.7742923 6 3 D 0.8 1 0.5560683
ggplot(newdata) + geom_violin(aes(y=Value, x=PH, color=HEALTH,fill='Model'), alpha=0.5)+ geom_violin(data=stehman, aes(y=BRATING,x=PH,color=HEALTH,fill='Obs'), alpha=0.5) + geom_point(data=stehman, aes(y=BRATING, x=PH, color=HEALTH), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=HEALTH, fill='Model', group=HEALTH, color=PH), alpha=0.5)+ geom_point(data=stehman, aes(y=BRATING, x=HEALTH, group=HEALTH,color=PH))

mcmc_intervals(as.matrix(stehman.brms), regex_pars='^b_|sigma')

mcmc_areas(as.matrix(stehman.brms), regex_pars='^b_|sigma')

- Explore parameter estimates
summary(stehman.mcmcpack)
Iterations = 1001:11000 Thinning interval = 1 Number of chains = 1 Sample size per chain = 10000 1. Empirical mean and standard deviation for each variable, plus standard error of the mean: Mean SD Naive SE Time-series SE (Intercept) 1.191647 0.10799 0.0010799 0.0010808 PH5.5 -0.383608 0.15268 0.0015268 0.0015402 PH7 -0.066463 0.15464 0.0015464 0.0015464 HEALTHH 0.424804 0.18944 0.0018944 0.0018944 PH5.5:HEALTHH -0.005671 0.29282 0.0029282 0.0029282 PH7:HEALTHH -0.210221 0.26962 0.0026962 0.0026962 sigma2 0.262782 0.04078 0.0004078 0.0004392 2. Quantiles for each variable: 2.5% 25% 50% 75% 97.5% (Intercept) 0.98291 1.1182 1.19201 1.26360 1.40429 PH5.5 -0.68239 -0.4863 -0.38285 -0.28048 -0.08895 PH7 -0.36713 -0.1721 -0.06551 0.03784 0.23973 HEALTHH 0.04734 0.2992 0.42443 0.55163 0.79540 PH5.5:HEALTHH -0.58371 -0.2030 -0.00225 0.19125 0.56058 PH7:HEALTHH -0.74732 -0.3911 -0.21045 -0.03046 0.32724 sigma2 0.19422 0.2337 0.25926 0.28757 0.35403#OR library(broom) tidyMCMC(stehman.mcmcpack,conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 (Intercept) 1.191646970 0.10798728 0.9749776 1.39536094 2 PH5.5 -0.383607638 0.15268450 -0.6840103 -0.09153912 3 PH7 -0.066462911 0.15464384 -0.3847130 0.21772860 4 HEALTHH 0.424804310 0.18943827 0.0502028 0.79684210 5 PH5.5:HEALTHH -0.005670638 0.29281702 -0.5780954 0.56501870 6 PH7:HEALTHH -0.210220999 0.26962165 -0.7621725 0.29989586 7 sigma2 0.262781583 0.04078248 0.1874536 0.34192612
#OR with p-values newdata = with(stehman, expand.grid(PH=levels(PH), HEALTH=levels(HEALTH))) Xmat = model.matrix(~PH*HEALTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(stehman.mcmcpack[,i]) )
[1] 0 [1] 0.0129 [1] 0.6676 [1] 0.0264 [1] 0.9871 [1] 0.4308
# Main effect of PH mcmcpvalue(stehman.mcmcpack[,which(wch==1)])
[1] 0.0289
# Main effect of HEALTH mcmcpvalue(stehman.mcmcpack[,which(wch==2)])
[1] 0.0264
# Interaction mcmcpvalue(stehman.mcmcpack[,which(wch==3)])
[1] 0.6808
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(BRATING~PH*HEALTH, data=stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435anova(lm(BRATING~PH*HEALTH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) PH 2 2.9293 1.46465 5.7099 0.004644 ** HEALTH 1 2.4273 2.42731 9.4629 0.002786 ** PH:HEALTH 2 0.1914 0.09569 0.3731 0.689691 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(BRATING~HEALTH*PH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) HEALTH 1 2.8910 2.89102 11.2706 0.00116 ** PH 2 2.4656 1.23280 4.8061 0.01042 * HEALTH:PH 2 0.1914 0.09569 0.3731 0.68969 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1print(stehman.r2jags)
Inference for Bugs model at "5", fit using jags, 3 chains, each with 50000 iterations (first 3000 discarded), n.thin = 10 n.sims = 14100 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 1.191 0.107 0.980 1.118 1.191 1.262 1.399 1.001 14000 beta[2] -0.381 0.151 -0.679 -0.483 -0.381 -0.280 -0.084 1.001 14000 beta[3] -0.067 0.155 -0.368 -0.171 -0.069 0.037 0.238 1.001 14000 beta[4] 0.427 0.191 0.057 0.297 0.426 0.555 0.808 1.001 5400 beta[5] -0.010 0.294 -0.582 -0.207 -0.007 0.186 0.570 1.001 10000 beta[6] -0.212 0.277 -0.754 -0.395 -0.211 -0.027 0.327 1.001 4200 sigma 0.514 0.040 0.445 0.486 0.512 0.539 0.600 1.001 12000 deviance 141.549 3.945 135.922 138.669 140.866 143.719 150.999 1.001 14000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 7.8 and DIC = 149.3 DIC is an estimate of expected predictive error (lower deviance is better).#OR library(broom) tidyMCMC(stehman.r2jags$BUGSoutput$sims.matrix,conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 beta[1] 1.19059004 0.10663274 0.97907535 1.3973732 2 beta[2] -0.38115261 0.15119821 -0.67413321 -0.0801445 3 beta[3] -0.06739250 0.15484579 -0.36270311 0.2416344 4 beta[4] 0.42725891 0.19092675 0.04528841 0.7936810 5 beta[5] -0.01003149 0.29430093 -0.60385438 0.5444966 6 beta[6] -0.21159909 0.27698287 -0.75890646 0.3197837 7 deviance 141.54892455 3.94526180 135.01465432 149.1233173 8 sigma 0.51442615 0.03974536 0.43921123 0.5926297
#OR with p-values newdata = with(stehman, expand.grid(PH=levels(PH), HEALTH=levels(HEALTH))) Xmat = model.matrix(~PH*HEALTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(stehman.r2jags$BUGSoutput$sims.matrix[,i]) )
[1] 0 [1] 0.01248227 [1] 0.6624823 [1] 0.02716312 [1] 0.9732624 [1] 0.4412057
# Main effect of PH mcmcpvalue(stehman.r2jags$BUGSoutput$sims.matrix[,which(wch==1)])
[1] 0.03049645
# Main effect of HEALTH mcmcpvalue(stehman.r2jags$BUGSoutput$sims.matrix[,which(wch==2)])
[1] 0.02716312
# Interaction mcmcpvalue(stehman.r2jags$BUGSoutput$sims.matrix[,which(wch==3)])
[1] 0.6978014
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(BRATING~PH*HEALTH, data=stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435anova(lm(BRATING~PH*HEALTH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) PH 2 2.9293 1.46465 5.7099 0.004644 ** HEALTH 1 2.4273 2.42731 9.4629 0.002786 ** PH:HEALTH 2 0.1914 0.09569 0.3731 0.689691 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(BRATING~HEALTH*PH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) HEALTH 1 2.8910 2.89102 11.2706 0.00116 ** PH 2 2.4656 1.23280 4.8061 0.01042 * HEALTH:PH 2 0.1914 0.09569 0.3731 0.68969 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1print(stehman.rstan, pars=c('beta','sigma'))
Inference for Stan model: fa2805f19ebd689dd8d4714ae0df073d. 3 chains, each with iter=2000; warmup=500; thin=3; post-warmup draws per chain=500, total post-warmup draws=1500. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 1.19 0.00 0.11 0.97 1.12 1.19 1.26 1.40 1304 1 beta[2] -0.38 0.00 0.15 -0.67 -0.49 -0.39 -0.28 -0.09 1374 1 beta[3] -0.07 0.00 0.16 -0.36 -0.18 -0.07 0.04 0.24 1330 1 beta[4] 0.42 0.01 0.19 0.04 0.30 0.42 0.54 0.78 1293 1 beta[5] 0.00 0.01 0.30 -0.59 -0.20 -0.01 0.20 0.58 1436 1 beta[6] -0.20 0.01 0.27 -0.72 -0.39 -0.20 -0.02 0.33 1204 1 sigma 0.51 0.00 0.04 0.44 0.49 0.51 0.54 0.60 1282 1 Samples were drawn using NUTS(diag_e) at Mon Dec 18 12:47:51 2017. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).#OR library(broom) tidyMCMC(stehman.rstan,conf.int=TRUE, conf.method='HPDinterval', pars=c('beta','sigma'))
term estimate std.error conf.low conf.high 1 beta[1] 1.193312651 0.10913406 0.98773252 1.40886288 2 beta[2] -0.383110829 0.15117033 -0.66129241 -0.08334005 3 beta[3] -0.069615802 0.15741971 -0.37745668 0.22885296 4 beta[4] 0.417077347 0.18886777 0.06756073 0.79669761 5 beta[5] -0.004156717 0.29749701 -0.62225074 0.55397148 6 beta[6] -0.201986470 0.27477591 -0.71897734 0.32990189 7 sigma 0.513884765 0.03913538 0.44151494 0.59540605
#OR with p-values newdata = with(stehman, expand.grid(PH=levels(PH), HEALTH=levels(HEALTH))) Xmat = model.matrix(~PH*HEALTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(stehman.rstan)[,i]) )
[1] 0 [1] 0.01066667 [1] 0.6666667 [1] 0.028 [1] 0.9906667 [1] 0.4553333
# Main effect of PH mcmcpvalue(as.matrix(stehman.rstan)[,which(wch==1)])
[1] 0.024
# Main effect of HEALTH mcmcpvalue(as.matrix(stehman.rstan)[,which(wch==2)])
[1] 0.028
# Interaction mcmcpvalue(as.matrix(stehman.rstan)[,which(wch==3)])
[1] 0.718
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(BRATING~PH*HEALTH, data=stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435anova(lm(BRATING~PH*HEALTH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) PH 2 2.9293 1.46465 5.7099 0.004644 ** HEALTH 1 2.4273 2.42731 9.4629 0.002786 ** PH:HEALTH 2 0.1914 0.09569 0.3731 0.689691 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(BRATING~HEALTH*PH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) HEALTH 1 2.8910 2.89102 11.2706 0.00116 ** PH 2 2.4656 1.23280 4.8061 0.01042 * HEALTH:PH 2 0.1914 0.09569 0.3731 0.68969 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(extract_log_lik(stehman.rstan)))
Computed from 1500 by 95 log-likelihood matrix Estimate SE elpd_loo -74.8 6.5 p_loo 7.3 1.3 looic 149.7 12.9 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.# now fit a model without main factor modelString=" data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } ransformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,1000); sigma~cauchy(0,5); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " Xmat <- model.matrix(~PH+HEALTH, stehman) stehman.list <- with(stehman,list(y=BRATING, X=Xmat,n=nrow(stehman), nX=ncol(Xmat))) stehman.rstan.red <- stan(data=stehman.list, model_code=modelString, chains=3, iter=2000, warmup=500, thin=3, refresh=FALSE )
Gradient evaluation took 2.9e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.29 seconds. Adjust your expectations accordingly! Elapsed Time: 0.028513 seconds (Warm-up) 0.090789 seconds (Sampling) 0.119302 seconds (Total) Gradient evaluation took 9e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds. Adjust your expectations accordingly! Elapsed Time: 0.028022 seconds (Warm-up) 0.075504 seconds (Sampling) 0.103526 seconds (Total) Gradient evaluation took 1.1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds. Adjust your expectations accordingly! Elapsed Time: 0.030623 seconds (Warm-up) 0.089719 seconds (Sampling) 0.120342 seconds (Total)(reduced=loo(extract_log_lik(stehman.rstan.red)))
Computed from 1500 by 95 log-likelihood matrix Estimate SE elpd_loo -72.6 6.4 p_loo 4.8 0.7 looic 145.2 12.7 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
summary(stehman.rstanarm)
Model Info: function: stan_glm family: gaussian [identity] formula: BRATING ~ PH * HEALTH algorithm: sampling priors: see help('prior_summary') sample: 2250 (posterior sample size) num obs: 95 Estimates: mean sd 2.5% 25% 50% 75% 97.5% (Intercept) 1.2 0.1 1.0 1.1 1.2 1.3 1.4 PH5.5 -0.4 0.1 -0.6 -0.5 -0.4 -0.3 -0.1 PH7 -0.1 0.1 -0.4 -0.2 -0.1 0.0 0.2 HEALTHH 0.4 0.2 0.1 0.3 0.4 0.5 0.7 PH5.5:HEALTHH 0.0 0.2 -0.5 -0.1 0.0 0.2 0.5 PH7:HEALTHH -0.1 0.2 -0.6 -0.3 -0.1 0.0 0.3 sigma 0.5 0.0 0.4 0.5 0.5 0.5 0.6 mean_PPD 1.2 0.1 1.0 1.1 1.2 1.2 1.3 log-posterior -80.6 1.9 -85.1 -81.6 -80.3 -79.2 -77.9 Diagnostics: mcse Rhat n_eff (Intercept) 0.0 1.0 1848 PH5.5 0.0 1.0 2005 PH7 0.0 1.0 1930 HEALTHH 0.0 1.0 1820 PH5.5:HEALTHH 0.0 1.0 1872 PH7:HEALTHH 0.0 1.0 1889 sigma 0.0 1.0 2110 mean_PPD 0.0 1.0 2053 log-posterior 0.0 1.0 1663 For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).#OR library(broom) tidyMCMC(stehman.rstanarm$stanfit,conf.int=TRUE, conf.method='HPDinterval', rhat=TRUE, ess=TRUE)
term estimate std.error conf.low conf.high rhat ess 1 (Intercept) 1.19367197 0.09811005 1.00267082 1.3794144 1.0015181 1848 2 PH5.5 -0.37103685 0.13716153 -0.62890963 -0.1015450 1.0005584 2005 3 PH7 -0.07738565 0.14394092 -0.36568565 0.2005097 1.0001321 1930 4 HEALTHH 0.38310472 0.16157074 0.05041117 0.6909403 0.9994089 1820 5 PH5.5:HEALTHH 0.01888488 0.24248171 -0.46984092 0.4773634 0.9995815 1872 6 PH7:HEALTHH -0.14853772 0.23370558 -0.58939364 0.3265783 0.9991106 1889 7 sigma 0.51176627 0.03809169 0.44268636 0.5919980 1.0004337 2110 8 mean_PPD 1.15056767 0.07384490 1.01347212 1.2986245 0.9999357 2053 9 log-posterior -80.60283406 1.86937990 -84.18945323 -77.4523925 0.9997297 1663
#OR with p-values newdata = with(stehman, expand.grid(PH=levels(PH), HEALTH=levels(HEALTH))) Xmat = model.matrix(~PH*HEALTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(stehman.rstanarm)[,i]) )
[1] 0 [1] 0.007111111 [1] 0.5982222 [1] 0.01822222 [1] 0.9431111 [1] 0.5244444
# Main effect of PH mcmcpvalue(as.matrix(stehman.rstanarm)[,which(wch==1)])
[1] 0.02
# Main effect of HEALTH mcmcpvalue(as.matrix(stehman.rstanarm)[,which(wch==2)])
[1] 0.01822222
# Interaction mcmcpvalue(as.matrix(stehman.rstanarm)[,which(wch==3)])
[1] 0.788
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(BRATING~PH*HEALTH, data=stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435anova(lm(BRATING~PH*HEALTH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) PH 2 2.9293 1.46465 5.7099 0.004644 ** HEALTH 1 2.4273 2.42731 9.4629 0.002786 ** PH:HEALTH 2 0.1914 0.09569 0.3731 0.689691 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(BRATING~HEALTH*PH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) HEALTH 1 2.8910 2.89102 11.2706 0.00116 ** PH 2 2.4656 1.23280 4.8061 0.01042 * HEALTH:PH 2 0.1914 0.09569 0.3731 0.68969 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(stehman.rstanarm))
Computed from 2250 by 95 log-likelihood matrix Estimate SE elpd_loo -73.9 6.4 p_loo 6.4 1.1 looic 147.9 12.8 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.stehman.rstanarm.red = update(stehman.rstanarm, .~PH+HEALTH)
Gradient evaluation took 4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.4 seconds. Adjust your expectations accordingly! Elapsed Time: 0.049038 seconds (Warm-up) 0.097841 seconds (Sampling) 0.146879 seconds (Total) Gradient evaluation took 1.9e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.19 seconds. Adjust your expectations accordingly! Elapsed Time: 0.044457 seconds (Warm-up) 0.100205 seconds (Sampling) 0.144662 seconds (Total) Gradient evaluation took 1.6e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.16 seconds. Adjust your expectations accordingly! Elapsed Time: 0.042975 seconds (Warm-up) 0.104433 seconds (Sampling) 0.147408 seconds (Total)(reduced=loo(stehman.rstanarm.red))
Computed from 2250 by 95 log-likelihood matrix Estimate SE elpd_loo -72.4 6.3 p_loo 4.6 0.7 looic 144.8 12.7 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
elpd_diff se 1.6 0.8There is very little support for an interaction (interaction effects small and loo of reduced model is smaller than the full model). There are main effectssummary(stehman.brms)
Family: gaussian(identity) Formula: BRATING ~ PH * HEALTH Data: stehman (Number of observations: 95) Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2; total post-warmup samples = 2250 ICs: LOO = NA; WAIC = NA; R2 = NA Population-Level Effects: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat Intercept 1.19 0.11 0.98 1.40 1807 1 PH5.5 -0.38 0.15 -0.67 -0.08 1731 1 PH7 -0.07 0.15 -0.38 0.22 1911 1 HEALTHH 0.42 0.18 0.06 0.78 1797 1 PH5.5:HEALTHH 0.01 0.28 -0.53 0.55 1392 1 PH7:HEALTHH -0.19 0.26 -0.69 0.33 1767 1 Family Specific Parameters: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat sigma 0.51 0.04 0.45 0.6 2144 1 Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat = 1).#OR library(broom) tidyMCMC(stehman.brms$fit,conf.int=TRUE, conf.method='HPDinterval', rhat=TRUE, ess=TRUE)
term estimate std.error conf.low conf.high rhat ess 1 b_Intercept 1.193674756 0.10530085 0.99311929 1.40494393 1.0003377 1807 2 b_PH5.5 -0.381747258 0.14726079 -0.68375335 -0.09339214 1.0002146 1731 3 b_PH7 -0.071524112 0.15253709 -0.38083958 0.21668439 0.9996752 1911 4 b_HEALTHH 0.417002902 0.18254053 0.06053687 0.78468311 0.9998761 1797 5 b_PH5.5:HEALTHH 0.007985665 0.27642096 -0.52552795 0.55176171 0.9999643 1392 6 b_PH7:HEALTHH -0.194003633 0.26341846 -0.69066979 0.32520555 0.9998097 1767 7 sigma 0.512449370 0.03793437 0.44254238 0.59053416 0.9988071 2144
#OR with p-values newdata = with(stehman, expand.grid(PH=levels(PH), HEALTH=levels(HEALTH))) Xmat = model.matrix(~PH*HEALTH, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(stehman.brms)[,i]) )
[1] 0 [1] 0.007555556 [1] 0.6337778 [1] 0.02533333 [1] 0.9782222 [1] 0.4586667
# Main effect of PH mcmcpvalue(as.matrix(stehman.brms)[,which(wch==1)])
[1] 0.02355556
# Main effect of HEALTH mcmcpvalue(as.matrix(stehman.brms)[,which(wch==2)])
[1] 0.02533333
# Interaction mcmcpvalue(as.matrix(stehman.brms)[,which(wch==3)])
[1] 0.6973333
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(BRATING~PH*HEALTH, data=stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435anova(lm(BRATING~PH*HEALTH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) PH 2 2.9293 1.46465 5.7099 0.004644 ** HEALTH 1 2.4273 2.42731 9.4629 0.002786 ** PH:HEALTH 2 0.1914 0.09569 0.3731 0.689691 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(BRATING~HEALTH*PH, data=stehman))
Analysis of Variance Table Response: BRATING Df Sum Sq Mean Sq F value Pr(>F) HEALTH 1 2.8910 2.89102 11.2706 0.00116 ** PH 2 2.4656 1.23280 4.8061 0.01042 * HEALTH:PH 2 0.1914 0.09569 0.3731 0.68969 Residuals 89 22.8293 0.25651 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(stehman.brms))
LOOIC SE 148.6 12.94
stehman.brms.red = update(stehman.brms, .~PH+HEALTH, refresh=FALSE)
Gradient evaluation took 2.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.25 seconds. Adjust your expectations accordingly! Elapsed Time: 0.046077 seconds (Warm-up) 0.04087 seconds (Sampling) 0.086947 seconds (Total) Gradient evaluation took 1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds. Adjust your expectations accordingly! Elapsed Time: 0.045096 seconds (Warm-up) 0.042914 seconds (Sampling) 0.08801 seconds (Total) Gradient evaluation took 1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds. Adjust your expectations accordingly! Elapsed Time: 0.045074 seconds (Warm-up) 0.042477 seconds (Sampling) 0.087551 seconds (Total)(reduced=loo(stehman.brms.red))
LOOIC SE 145.15 12.69
par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
- Explore the general effect of HEALTH across all PHs
mcmc = stehman.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^PH|^HEALTH", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 0.3528404 0.1168168 0.1267973 0.5815175
# OR if we express this as a percentage change Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 34.36308 12.51254 10.38557 58.69549
mcmc = stehman.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 0.353382 0.1182966 0.1269179 0.58607
# OR if we express this as a percentage change Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 34.43986 12.69745 11.23016 60.72792
mcmc = as.matrix(stehman.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 0.348363 0.1219291 0.1075704 0.5740102
# OR if we express this as a percentage change Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 33.95054 13.13565 9.121375 59.18455
mcmc = as.matrix(stehman.rstanarm) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^PH|^HEALTH',colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 0.3398871 0.112829 0.1278381 0.5567404
# OR if we express this as a percentage change Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 33.00564 12.0627 10.93289 56.97729
mcmc = as.matrix(stehman.brms) wch = grep('^b_',colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] Xmat = Xmat[2,] - Xmat[1,] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 2 0.3549969 0.1147896 0.1205326 0.5633382
# OR if we express this as a percentage change Xmat = model.matrix(~PH*HEALTH,newdata) Xmat = aggregate(Xmat, list(newdata$HEALTH),FUN=mean)[,-1] coefs = mcmc[,wch] fit=coefs %*% t(Xmat) fit = 100*(fit[,2] - fit[,1])/fit[,1] tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 var1 34.50187 12.24638 10.6126 58.24672
- Generate a summary figure
mcmc = stehman.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^PH|^HEALTH", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
PH HEALTH estimate std.error conf.low conf.high 1 3 D 1.1916470 0.1079873 0.9749776 1.395361 2 5.5 D 0.8080393 0.1068135 0.6016267 1.021239 3 7 D 1.1251841 0.1119449 0.8989564 1.331882 4 3 H 1.6164513 0.1554543 1.3075314 1.921243 5 5.5 H 1.2271730 0.1941066 0.8237557 1.590138 6 7 H 1.3397674 0.1629537 1.0142307 1.648695
ggplot(newdata, aes(y=estimate, x=PH, fill=HEALTH)) + geom_blank() + geom_line(aes(x=as.numeric(PH), linetype=HEALTH), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=HEALTH), size=3, position=position_dodge(width=0.2))+ scale_y_continuous('Bud emergence rating')+ scale_x_discrete('PH')+ scale_shape_manual('HEALTH',values=c(21,16))+ scale_fill_manual('HEALTH',values=c('white','black'))+ scale_linetype_manual('HEALTH',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

mcmc = stehman.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
PH HEALTH estimate std.error conf.low conf.high 1 3 D 1.1905900 0.1066327 0.9790754 1.397373 2 5.5 D 0.8094374 0.1078854 0.5964688 1.018619 3 7 D 1.1231975 0.1119311 0.9115395 1.349675 4 3 H 1.6178489 0.1565902 1.3285247 1.936248 5 5.5 H 1.2266648 0.1966736 0.8470403 1.616027 6 7 H 1.3388574 0.1642197 1.0122264 1.661033
ggplot(newdata, aes(y=estimate, x=PH, fill=HEALTH)) + geom_blank() + geom_line(aes(x=as.numeric(PH), linetype=HEALTH), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high, position=position_dodge(width=0.2)))+ geom_point(aes(shape=HEALTH), size=3, position=position_dodge(width=0.2))+ scale_y_continuous('Bud emergence rating')+ scale_x_discrete('PH')+ scale_shape_manual('HEALTH',values=c(21,16))+ scale_fill_manual('HEALTH',values=c('white','black'))+ scale_linetype_manual('HEALTH',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))
Error: Aesthetics must be either length 1 or the same as the data (6): ymin, ymax, position, x, y, fill

mcmc = as.matrix(stehman.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) Xmat = model.matrix(~PH*HEALTH,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
PH HEALTH estimate std.error conf.low conf.high 1 3 D 1.1933127 0.1091341 0.9877325 1.408863 2 5.5 D 0.8102018 0.1074251 0.5913518 1.020503 3 7 D 1.1236968 0.1153893 0.9007333 1.348498 4 3 H 1.6103900 0.1520670 1.3336927 1.924424 5 5.5 H 1.2231225 0.1997801 0.8717571 1.667370 6 7 H 1.3387877 0.1676729 1.0148635 1.671485
ggplot(newdata, aes(y=estimate, x=PH, fill=HEALTH)) + geom_blank() + geom_line(aes(x=as.numeric(PH), linetype=HEALTH), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=HEALTH), size=3, position=position_dodge(width=0.2))+ scale_y_continuous('Bud emergence rating')+ scale_x_discrete('PH')+ scale_shape_manual('HEALTH',values=c(21,16))+ scale_fill_manual('HEALTH',values=c('white','black'))+ scale_linetype_manual('HEALTH',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

newdata = expand.grid(PH=levels(stehman$PH), HEALTH=levels(stehman$HEALTH)) fit = posterior_linpred(stehman.rstanarm, newdata = newdata) newdata = newdata %>% cbind(tidyMCMC(as.mcmc(fit),conf.int = TRUE,conf.method = "HPDinterval")) ggplot(newdata, aes(y=estimate, x=PH, fill=HEALTH)) + geom_blank() + geom_line(aes(x=as.numeric(PH), linetype=HEALTH), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=HEALTH), size=3, position=position_dodge(width=0.2))+ scale_y_continuous('Bud emergence rating')+ scale_x_discrete('PH')+ scale_shape_manual('HEALTH',values=c(21,16))+ scale_fill_manual('HEALTH',values=c('white','black'))+ scale_linetype_manual('HEALTH',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

## The simple way plot(marginal_effects(stehman.brms))



## OR eff=marginal_effects(stehman.brms) ggplot(eff[['PH:HEALTH']], aes(y=estimate__, x=PH, fill=HEALTH)) + geom_blank() + geom_line(aes(x=as.numeric(PH), linetype=HEALTH), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=lower__, ymax=upper__), position=position_dodge(width=0.2))+ geom_point(aes(shape=HEALTH), size=3, position=position_dodge(width=0.2))+ scale_y_continuous('Bud emergence rating')+ scale_x_discrete('PH')+ scale_shape_manual('HEALTH',values=c(21,16))+ scale_fill_manual('HEALTH',values=c('white','black'))+ scale_linetype_manual('HEALTH',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

- Explore finite-population standard deviations
mcmc = stehman.mcmcpack Xmat = model.matrix(~PH*HEALTH, stehman) wch = attr(Xmat, 'assign') wch
[1] 0 1 1 2 3 3
# Get the rowwise standard deviations between effects parameters sd.PH = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.HEALTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^PH|^HEALTH',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,stehman$BRATING,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.PH, sd.HEALTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 0.2203599 0.072962160 0.07833344 0.3628514 2 sd.HEALTH 0.3015136 0.131385872 0.03549874 0.5517216 3 sd.Int 0.1573470 0.080911095 0.01668094 0.3106556 4 sd.resid 0.5068045 0.009086833 0.49356407 0.5242985
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 18.84990 6.089890 6.721980 30.07697 2 sd.HEALTH 25.91514 8.593741 6.605029 39.71880 3 sd.Int 12.73531 5.409516 2.639222 22.96541 4 sd.resid 43.33362 6.354587 31.992006 56.13694
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = stehman.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~PH*HEALTH, stehman) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.PH = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.HEALTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,stehman$BRATING,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.PH, sd.HEALTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 0.2189424 0.072552621 0.07354368 0.3580210 2 sd.HEALTH 0.3033256 0.132269116 0.04026688 0.5558738 3 sd.Int 0.1590139 0.082487683 0.01113549 0.3111126 4 sd.resid 0.5069435 0.009124031 0.49358461 0.5249415
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 18.61379 6.047225 6.988233 30.39436 2 sd.HEALTH 26.07474 8.527034 6.575304 39.42712 3 sd.Int 12.92043 5.452122 2.606432 23.18682 4 sd.resid 43.24714 6.451129 31.182928 55.88777
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(stehman.rstan) Xmat = model.matrix(~PH*HEALTH, stehman) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.PH = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.HEALTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,stehman$BRATING,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.PH, sd.HEALTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 0.2199985 0.071772512 0.08048919 0.3529833 2 sd.HEALTH 0.2967786 0.129360018 0.05090361 0.5523131 3 sd.Int 0.1579971 0.082172719 0.02195667 0.3165202 4 sd.resid 0.5070408 0.008951356 0.49428764 0.5246637
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 18.94900 5.990250 6.567436 29.60071 2 sd.HEALTH 25.76607 8.473636 5.542474 37.46864 3 sd.Int 12.75490 5.471206 2.797328 23.12735 4 sd.resid 43.50229 6.426886 32.811249 57.35298
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(stehman.rstanarm) Xmat = model.matrix(~PH*HEALTH, stehman) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.PH = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.HEALTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^PH|^HEALTH',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,stehman$BRATING,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.PH, sd.HEALTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 0.2101966 0.066962799 0.07608042 0.3378495 2 sd.HEALTH 0.2711877 0.113553187 0.03711287 0.4885686 3 sd.Int 0.1311117 0.065965411 0.01505872 0.2595815 4 sd.resid 0.5053570 0.007890909 0.49379887 0.5206185
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 18.96698 5.643682 7.468847 29.70428 2 sd.HEALTH 24.55119 8.022345 7.747575 39.66649 3 sd.Int 11.28375 4.894101 2.070110 20.41055 4 sd.resid 45.46272 6.389440 33.941426 58.14420
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(stehman.brms) Xmat = model.matrix(~PH*HEALTH, stehman) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.PH = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.HEALTH = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^b_',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,stehman$BRATING,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.PH, sd.HEALTH, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 0.2182540 0.069998050 0.08198533 0.3633113 2 sd.HEALTH 0.2961006 0.126215484 0.03830631 0.5347764 3 sd.Int 0.1504554 0.076861825 0.01736893 0.2951451 4 sd.resid 0.5061225 0.008700461 0.49333325 0.5221924
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.PH 18.93863 5.847734 7.277636 30.44363 2 sd.HEALTH 25.72749 8.363631 7.204982 39.20808 3 sd.Int 12.38404 5.253651 1.787843 21.87904 4 sd.resid 43.76098 6.346921 32.083012 56.62989
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

- Estimate a psuedo-$R^2$
library(broom) mcmc <- stehman.mcmcpack Xmat = model.matrix(~PH*HEALTH, stehman) wch = c(which(colnames(mcmc)=='(Intercept)'), grep('^PH|^HEALTH', colnames(mcmc))) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, stehman$BRATING, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.2158761 0.06095225 0.09671562 0.3321677
#for comparison with frequentist summary(lm(BRATING ~ PH*HEALTH, stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435library(broom) mcmc <- stehman.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~PH*HEALTH, stehman) wch = grep('^beta', colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, stehman$BRATING, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.2157242 0.06133794 0.09880009 0.3345779
#for comparison with frequentist summary(lm(BRATING ~ PH*HEALTH, stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435library(broom) mcmc <- as.matrix(stehman.rstan) Xmat = model.matrix(~PH*HEALTH, stehman) wch = grep('^beta', colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, stehman$BRATING, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.2137876 0.06132725 0.09694539 0.3316519
#for comparison with frequentist summary(lm(BRATING ~ PH*HEALTH, stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435library(broom) mcmc <- as.matrix(stehman.rstanarm) Xmat = model.matrix(~PH*HEALTH, stehman) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^PH|^HEALTH',colnames(mcmc))) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, stehman$BRATING, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.1988162 0.06015237 0.0820395 0.3169533
#for comparison with frequentist summary(lm(BRATING ~ PH*HEALTH, stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435library(broom) mcmc <- as.matrix(stehman.brms) Xmat = model.matrix(~PH*HEALTH, stehman) wch = grep('^b_',colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, stehman$BRATING, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.2128962 0.06024065 0.1018493 0.3360714
#for comparison with frequentist summary(lm(BRATING ~ PH*HEALTH, stehman))
Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.191304 0.105606 11.281 <2e-16 *** PH5.5 -0.382609 0.149349 -2.562 0.0121 * PH7 -0.067495 0.152863 -0.442 0.6599 HEALTHH 0.426877 0.185665 2.299 0.0238 * PH5.5:HEALTHH -0.007002 0.286824 -0.024 0.9806 PH7:HEALTHH -0.210687 0.268956 -0.783 0.4355 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.5065 on 89 degrees of freedom Multiple R-squared: 0.1955, Adjusted R-squared: 0.1503 F-statistic: 4.326 on 5 and 89 DF, p-value: 0.001435
Two-factor ANOVA with substantial interactions
An ecologist studying a rocky shore at Phillip Island, in southeastern Australia, was interested in how clumps of intertidal mussels are maintained. In particular, he wanted to know how densities of adult mussels affected recruitment of young individuals from the plankton. As with most marine invertebrates, recruitment is highly patchy in time, so he expected to find seasonal variation, and the interaction between season and density - whether effects of adult mussel density vary across seasons - was the aspect of most interest.
The data were collected from four seasons, and with two densities of adult mussels. The experiment consisted of clumps of adult mussels attached to the rocks. These clumps were then brought back to the laboratory, and the number of baby mussels recorded. There were 3-6 replicate clumps for each density and season combination.
Download Quinn data set| Format of quinn.csv data files | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
quinn <- read.table("../downloads/data/quinn.csv", header = T, sep = ",", strip.white = T) head(quinn)
SEASON DENSITY RECRUITS SQRTRECRUITS GROUP 1 Spring Low 15 3.872983 SpringLow 2 Spring Low 10 3.162278 SpringLow 3 Spring Low 13 3.605551 SpringLow 4 Spring Low 13 3.605551 SpringLow 5 Spring Low 5 2.236068 SpringLow 6 Spring High 11 3.316625 SpringHigh
Exploratory data analysis suggested that the response variable (RECRUITS) was not normally distributed. This variable represents the observed number of newly recruited mussels. As a count, it is likely that the underlying process is a Poisson process rather than Gaussian. Quinn elected to normalize the number of recruits by applying a square-root transformation. Such a transformation was chosen over a logarithmic transformation due to the presence of zeros.
Whilst applying root transformations was a reasonably common practice for addressing non-normality in count data, it does have some very undesirable consequences. When back-transforming predictions (and effects) into the natural scale, it is important to remember that the inverse of a root transformation is not monotonic (that is, order is not preserved over the entire range of possible values). Consider back-tranforming from the following ordered sequence: -4,0.5,0.8,2. The back-transforms would be: 16,0.25,0.64,4.
In a later tutorial we will revisit this example and more appropriately fit the model against a Poisson and Negative Binomial distributions. However, for the current example, we will apply the square-root transformation so as to see the impacts of such a action. Note, there is already a variable in the dataset called SQRTRECRUITS. For some modelling routines there is an advantage to performing the transformations inline (this way the transformation is captured in the model call and therefore induce automatic back-transformations). However, this is not the case for Bayesian routines and it is arguably better to use a new transformed variable (if at all).
- Fit the model to investigate the effects of season and adult density on the
recruitment (square-root transformed) of mussels.
$$
\begin{align}
y_{ij} &\sim{} N(\mu_{ij}, \sigma)\\
\mu_{ij} &= \beta_0 + \mathbf{X}\boldsymbol{\beta}\\[1em]
\beta_0 &\sim{} N(0,10)\\
\beta &\sim{} N(0,1)\\
\sigma &\sim{} cauchy(0,2)\\
\end{align}
$$
library(MCMCpack) quinn.mcmcpack = MCMCregress(SQRTRECRUITS ~ SEASON * DENSITY, data = quinn)
modelString = " model { #Likelihood for (i in 1:n) { y[i]~dnorm(mu[i],tau) mu[i] <- inprod(beta[],X[i,]) } #Priors beta0 ~ dnorm(0.01,1.0E-6) for (j in 1:nX) { beta[j] ~ dnorm(0.01,1.0E-6) } tau <- 1 / (sigma * sigma) sigma~dunif(0,100) } " X = model.matrix(~SEASON * DENSITY, data = quinn) quinn.list <- with(quinn, list(y = SQRTRECRUITS, X = X, nX = ncol(X), n = nrow(quinn))) params <- c("beta", "sigma") burnInSteps = 3000 nChains = 3 numSavedSteps = 15000 thinSteps = 10 nIter = ceiling((numSavedSteps * thinSteps)/nChains) quinn.r2jags <- jags(data = quinn.list, inits = NULL, parameters.to.save = params, model.file = textConnection(modelString), n.chains = nChains, n.iter = nIter, n.burnin = burnInSteps, n.thin = thinSteps)
Compiling model graph Resolving undeclared variables Allocating nodes Graph information: Observed stochastic nodes: 42 Unobserved stochastic nodes: 10 Total graph size: 464 Initializing model
modelString = " data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } ransformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,10); sigma~cauchy(0,2); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " X = model.matrix(~SEASON * DENSITY, data = quinn) quinn.list <- with(quinn, list(y = SQRTRECRUITS, X = X, nX = ncol(X), n = nrow(quinn))) quinn.rstan <- stan(data = quinn.list, model_code = modelString, chains = 3, iter = 2000, warmup = 500, thin = 3)
In file included from /usr/local/lib/R/site-library/BH/include/boost/config.hpp:39:0, from /usr/local/lib/R/site-library/BH/include/boost/math/tools/config.hpp:13, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/var.hpp:7, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/gevv_vvv_vari.hpp:5, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core.hpp:12, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/mat.hpp:4, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math.hpp:4, from /usr/local/lib/R/site-library/StanHeaders/include/src/stan/model/model_header.hpp:4, from file574125ed90ff.cpp:8: /usr/local/lib/R/site-library/BH/include/boost/config/compiler/gcc.hpp:186:0: warning: "BOOST_NO_CXX11_RVALUE_REFERENCES" redefined # define BOOST_NO_CXX11_RVALUE_REFERENCES ^ <command-line>:0:0: note: this is the location of the previous definition SAMPLING FOR MODEL 'a6ade6a40138b983c8773faeb7752a0c' NOW (CHAIN 1). Gradient evaluation took 2.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.24 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.05226 seconds (Warm-up) 0.139186 seconds (Sampling) 0.191446 seconds (Total) SAMPLING FOR MODEL 'a6ade6a40138b983c8773faeb7752a0c' NOW (CHAIN 2). Gradient evaluation took 9e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.047917 seconds (Warm-up) 0.106509 seconds (Sampling) 0.154426 seconds (Total) SAMPLING FOR MODEL 'a6ade6a40138b983c8773faeb7752a0c' NOW (CHAIN 3). Gradient evaluation took 8e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.08 seconds. Adjust your expectations accordingly! Iteration: 1 / 2000 [ 0%] (Warmup) Iteration: 200 / 2000 [ 10%] (Warmup) Iteration: 400 / 2000 [ 20%] (Warmup) Iteration: 501 / 2000 [ 25%] (Sampling) Iteration: 700 / 2000 [ 35%] (Sampling) Iteration: 900 / 2000 [ 45%] (Sampling) Iteration: 1100 / 2000 [ 55%] (Sampling) Iteration: 1300 / 2000 [ 65%] (Sampling) Iteration: 1500 / 2000 [ 75%] (Sampling) Iteration: 1700 / 2000 [ 85%] (Sampling) Iteration: 1900 / 2000 [ 95%] (Sampling) Iteration: 2000 / 2000 [100%] (Sampling) Elapsed Time: 0.0459 seconds (Warm-up) 0.114115 seconds (Sampling) 0.160015 seconds (Total)print(quinn.rstan, par = c("beta", "sigma"))
Inference for Stan model: a6ade6a40138b983c8773faeb7752a0c. 3 chains, each with iter=2000; warmup=500; thin=3; post-warmup draws per chain=500, total post-warmup draws=1500. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 4.25 0.01 0.43 3.37 3.97 4.25 4.53 5.05 1122 1 beta[2] -1.24 0.02 0.59 -2.44 -1.63 -1.22 -0.83 -0.15 1023 1 beta[3] 2.61 0.02 0.60 1.38 2.21 2.60 3.01 3.86 788 1 beta[4] -1.98 0.02 0.61 -3.16 -2.38 -1.99 -1.57 -0.76 1062 1 beta[5] -0.01 0.02 0.66 -1.30 -0.45 -0.03 0.42 1.30 1007 1 beta[6] 0.29 0.03 0.92 -1.50 -0.30 0.28 0.90 2.10 1067 1 beta[7] -2.19 0.03 0.90 -3.93 -2.77 -2.19 -1.59 -0.45 1107 1 beta[8] -1.31 0.03 0.97 -3.19 -1.95 -1.33 -0.64 0.56 1270 1 sigma 1.04 0.00 0.13 0.83 0.95 1.03 1.11 1.32 1473 1 Samples were drawn using NUTS(diag_e) at Tue Dec 19 08:18:10 2017. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).quinn.rstanarm = stan_glm(SQRTRECRUITS ~ SEASON * DENSITY, data = quinn, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior_intercept = normal(0, 10), prior = normal(0, 1), prior_aux = cauchy(0, 2))
Gradient evaluation took 6.2e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.62 seconds. Adjust your expectations accordingly! Elapsed Time: 0.072712 seconds (Warm-up) 0.153801 seconds (Sampling) 0.226513 seconds (Total) Gradient evaluation took 1.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds. Adjust your expectations accordingly! Elapsed Time: 0.057856 seconds (Warm-up) 0.147768 seconds (Sampling) 0.205624 seconds (Total) Gradient evaluation took 1.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds. Adjust your expectations accordingly! Elapsed Time: 0.059313 seconds (Warm-up) 0.165404 seconds (Sampling) 0.224717 seconds (Total)print(quinn.rstanarm)
stan_glm family: gaussian [identity] formula: SQRTRECRUITS ~ SEASON * DENSITY ------ Estimates: Median MAD_SD (Intercept) 4.3 0.4 SEASONSpring -1.2 0.5 SEASONSummer 2.4 0.5 SEASONWinter -2.0 0.5 DENSITYLow -0.2 0.5 SEASONSpring:DENSITYLow 0.4 0.8 SEASONSummer:DENSITYLow -1.7 0.7 SEASONWinter:DENSITYLow -1.0 0.8 sigma 1.0 0.1 Sample avg. posterior predictive distribution of y (X = xbar): Median MAD_SD mean_PPD 3.9 0.2 ------ For info on the priors used see help('prior_summary.stanreg').tidyMCMC(quinn.rstanarm, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 (Intercept) 4.2944621 0.3781257 3.6008854 5.0780981 2 SEASONSpring -1.1944391 0.5415104 -2.2425511 -0.1488528 3 SEASONSummer 2.3506163 0.5418177 1.2919129 3.4111662 4 SEASONWinter -1.9593821 0.5441314 -3.0178154 -0.8888779 5 DENSITYLow -0.2031083 0.5393479 -1.2823813 0.8410013 6 SEASONSpring:DENSITYLow 0.3687119 0.7663820 -1.1687244 1.8007469 7 SEASONSummer:DENSITYLow -1.6934501 0.7516600 -3.2933392 -0.2561633 8 SEASONWinter:DENSITYLow -1.0512296 0.8304569 -2.6875590 0.4954263 9 sigma 1.0430770 0.1315636 0.8032712 1.2960622
quinn.brms = brm(SQRTRECRUITS ~ SEASON * DENSITY, data = quinn, iter = 2000, warmup = 500, chains = 3, thin = 2, refresh = 0, prior = c(prior(normal(0, 10), class = "Intercept"), prior(normal(0, 1), class = "b"), prior(cauchy(0, 2), class = "sigma")))
Gradient evaluation took 1.8e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.18 seconds. Adjust your expectations accordingly! Elapsed Time: 0.035472 seconds (Warm-up) 0.083562 seconds (Sampling) 0.119034 seconds (Total) Gradient evaluation took 9e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds. Adjust your expectations accordingly! Elapsed Time: 0.036017 seconds (Warm-up) 0.089386 seconds (Sampling) 0.125403 seconds (Total) Gradient evaluation took 1e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds. Adjust your expectations accordingly! Elapsed Time: 0.033439 seconds (Warm-up) 0.080357 seconds (Sampling) 0.113796 seconds (Total)print(quinn.brms)
Family: gaussian(identity) Formula: SQRTRECRUITS ~ SEASON * DENSITY Data: quinn (Number of observations: 42) Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2; total post-warmup samples = 2250 ICs: LOO = NA; WAIC = NA; R2 = NA Population-Level Effects: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat Intercept 4.29 0.33 3.63 4.93 1854 1 SEASONSpring -1.02 0.46 -1.91 -0.07 1884 1 SEASONSummer 2.00 0.48 1.02 2.92 1887 1 SEASONWinter -1.83 0.46 -2.75 -0.95 1941 1 DENSITYLow -0.37 0.42 -1.19 0.41 1890 1 SEASONSpring:DENSITYLow 0.31 0.60 -0.87 1.50 1962 1 SEASONSummer:DENSITYLow -1.07 0.60 -2.19 0.15 1979 1 SEASONWinter:DENSITYLow -0.85 0.65 -2.15 0.34 1939 1 Family Specific Parameters: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat sigma 1.06 0.14 0.83 1.36 1776 1 Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat = 1).tidyMCMC(quinn.brms, conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 b_Intercept 4.2937368 0.3267812 3.6208843 4.92899297 2 b_SEASONSpring -1.0204416 0.4627238 -1.8869003 -0.05501539 3 b_SEASONSummer 1.9975818 0.4765084 1.1050091 2.99362386 4 b_SEASONWinter -1.8325778 0.4591678 -2.6768481 -0.88445027 5 b_DENSITYLow -0.3696387 0.4230952 -1.2161672 0.38096496 6 b_SEASONSpring:DENSITYLow 0.3098758 0.5999900 -0.9051438 1.44819946 7 b_SEASONSummer:DENSITYLow -1.0658827 0.6002369 -2.2027007 0.12982398 8 b_SEASONWinter:DENSITYLow -0.8468540 0.6453871 -2.1293147 0.35870271 9 sigma 1.0602483 0.1361341 0.8138045 1.33499319
- Explore MCMC diagnostics
library(MCMCpack) plot(quinn.mcmcpack)



raftery.diag(quinn.mcmcpack)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) (Intercept) 2 3802 3746 1.010 SEASONSpring 2 3819 3746 1.020 SEASONSummer 2 3851 3746 1.030 SEASONWinter 2 3851 3746 1.030 DENSITYLow 2 3834 3746 1.020 SEASONSpring:DENSITYLow 2 3802 3746 1.010 SEASONSummer:DENSITYLow 2 3741 3746 0.999 SEASONWinter:DENSITYLow 2 3772 3746 1.010 sigma2 2 3865 3746 1.030autocorr.diag(quinn.mcmcpack)
(Intercept) SEASONSpring SEASONSummer SEASONWinter DENSITYLow SEASONSpring:DENSITYLow Lag 0 1.000000000 1.000000000 1.00000000 1.000000000 1.000000000 1.000000000 Lag 1 -0.007643649 -0.026132560 0.00321781 0.005202947 -0.011808793 -0.024459106 Lag 5 -0.015081799 -0.008048516 -0.00920160 -0.008282817 0.001280582 -0.006105166 Lag 10 0.014313873 0.012699385 0.01209427 -0.007232773 -0.009890837 0.015217312 Lag 50 0.010836422 0.003125721 0.01994357 0.006052357 0.006596454 0.014338348 SEASONSummer:DENSITYLow SEASONWinter:DENSITYLow sigma2 Lag 0 1.000000000 1.000000000 1.000000000 Lag 1 0.005026989 -0.014317948 0.194845073 Lag 5 0.016458386 -0.020171068 -0.002756425 Lag 10 -0.016776058 -0.010438622 -0.004218901 Lag 50 -0.006786834 -0.001237867 0.013569038quinn.mcmc = as.mcmc(quinn.r2jags) plot(quinn.mcmc)




preds <- grep("beta", colnames(quinn.mcmc[[1]])) plot(quinn.mcmc[, preds])


raftery.diag(quinn.mcmc)
[[1]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 10 37660 3746 10.10 beta[2] 20 38330 3746 10.20 beta[3] 20 38330 3746 10.20 beta[4] 20 35750 3746 9.54 beta[5] 20 39000 3746 10.40 beta[6] 20 38330 3746 10.20 beta[7] 30 41100 3746 11.00 beta[8] 10 37660 3746 10.10 deviance 10 37660 3746 10.10 sigma 10 37660 3746 10.10 [[2]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 10 37660 3746 10.10 beta[2] 20 38350 3746 10.20 beta[3] 20 38330 3746 10.20 beta[4] 20 36380 3746 9.71 beta[5] 20 38330 3746 10.20 beta[6] 20 39000 3746 10.40 beta[7] 20 37020 3746 9.88 beta[8] 20 36380 3746 9.71 deviance 10 37660 3746 10.10 sigma 20 37020 3746 9.88 [[3]] Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 Burn-in Total Lower bound Dependence (M) (N) (Nmin) factor (I) beta[1] 20 35750 3746 9.54 beta[2] 20 35750 3746 9.54 beta[3] 20 37020 3746 9.88 beta[4] 20 36380 3746 9.71 beta[5] 10 37660 3746 10.10 beta[6] 20 39680 3746 10.60 beta[7] 10 37660 3746 10.10 beta[8] 20 37020 3746 9.88 deviance 20 38330 3746 10.20 sigma 20 35750 3746 9.54autocorr.diag(quinn.mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] Lag 0 1.0000000000 1.0000000000 1.000000000 1.000000000 1.0000000000 1.000000000 Lag 10 0.0028152468 -0.0080705893 -0.001292731 -0.006689358 0.0091533386 0.002714831 Lag 50 -0.0045311158 -0.0089963411 -0.012837348 -0.001202299 -0.0004072487 -0.005378027 Lag 100 0.0007329071 -0.0002741862 -0.006573026 0.001477677 0.0129954819 -0.010727888 Lag 500 0.0057890673 -0.0028312361 -0.004841524 0.001352826 -0.0006000175 -0.010883766 beta[7] beta[8] deviance sigma Lag 0 1.0000000000 1.000000000 1.000000000 1.00000000 Lag 10 0.0077476020 -0.010767691 -0.002040765 -0.01097413 Lag 50 -0.0065906254 0.002997587 -0.006455403 -0.01064747 Lag 100 0.0053403043 0.006990484 -0.005939086 0.00378940 Lag 500 -0.0001268999 0.003454704 0.005335681 -0.00179597s = as.array(quinn.rstan) wch = grep("beta", dimnames(s)$parameters) s = s[, , wch] mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)


autocorr.diag(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] Lag 0 1.000000000 1.000000000 1.000000000 1.000000000 1.000000000 1.00000000 1.00000000 Lag 1 0.125589322 0.098988597 0.144890737 0.117213132 0.101184364 0.06409708 0.09902766 Lag 5 0.012841832 0.044506310 0.057083754 -0.005310630 0.025183096 0.05521445 0.04947603 Lag 10 -0.002871466 -0.038114462 -0.009460004 0.003268104 -0.004004047 0.01794970 -0.02022037 Lag 50 -0.024011789 -0.009202461 0.025768387 0.001594138 0.020423207 0.01723572 0.03294167
## Or via rstan stan_trace(quinn.rstan)

stan_ac(quinn.rstan)

stan_rhat(quinn.rstan)

stan_ess(quinn.rstan)

## Or via bayesplot detach("package:reshape") mcmc_trace(as.matrix(quinn.rstan), regex_pars = "beta|sigma")

mcmc_dens(as.matrix(quinn.rstan), regex_pars = "beta|sigma")

s = as.array(quinn.rstanarm) mcmc <- do.call(mcmc.list, plyr:::alply(s[, , -(length(s[1, 1, ]))], 2, as.mcmc)) plot(mcmc)


autocorr.diag(mcmc)
(Intercept) SEASONSpring SEASONSummer SEASONWinter DENSITYLow SEASONSpring:DENSITYLow Lag 0 1.00000000 1.000000000 1.0000000000 1.000000000 1.000000000 1.000000000 Lag 1 0.12402868 0.073460288 0.1080337237 0.084533939 0.163721063 0.116710530 Lag 5 -0.01426218 -0.009058940 -0.0004648023 -0.015061355 -0.024215498 -0.041218597 Lag 10 0.01709041 0.007013523 -0.0497780688 0.005669201 -0.003557755 0.011361252 Lag 50 0.01969934 0.042097184 0.0178208806 -0.035452694 0.005219093 -0.006616679 SEASONSummer:DENSITYLow SEASONWinter:DENSITYLow Lag 0 1.00000000 1.00000000 Lag 1 0.14738795 0.10196544 Lag 5 -0.03053775 -0.01956079 Lag 10 -0.02629748 -0.02175141 Lag 50 0.01467429 -0.01873360## OR via rstan stan_trace(quinn.rstanarm)

raftery.diag(quinn.rstanarm)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
stan_ac(quinn.rstanarm)

stan_rhat(quinn.rstanarm)

stan_ess(quinn.rstanarm)

## OR via bayesplot detach("package:reshape") mcmc_trace(as.array(quinn.rstanarm), regex_pars = "Intercept|SEASON|DENSITY|sigma")

mcmc_dens(as.array(quinn.rstanarm))

posterior_vs_prior(quinn.rstanarm, color_by = "vs", group_by = TRUE, facet_args = list(scales = "free_y"))
Gradient evaluation took 3.9e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.39 seconds. Adjust your expectations accordingly! Elapsed Time: 0.039845 seconds (Warm-up) 0.112468 seconds (Sampling) 0.152313 seconds (Total) Gradient evaluation took 1.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds. Adjust your expectations accordingly! Elapsed Time: 0.042003 seconds (Warm-up) 0.115089 seconds (Sampling) 0.157092 seconds (Total)
mcmc = as.mcmc(quinn.brms) plot(mcmc)




autocorr.diag(mcmc)
Error in ts(x, start = start(x), end = end(x), deltat = thin(x)): invalid time series parameters specified
## OR via rstan stan_trace(quinn.brms$fit)

raftery.diag(quinn.brms)
Quantile (q) = 0.025 Accuracy (r) = +/- 0.005 Probability (s) = 0.95 You need a sample size of at least 3746 with these values of q, r and s
stan_ac(quinn.brms$fit)

stan_rhat(quinn.brms$fit)

stan_ess(quinn.brms$fit)

- Explore model validation
mcmc = as.data.frame(quinn.mcmcpack) #generate a model matrix newdata = quinn Xmat = model.matrix(~SEASON*DENSITY, newdata) ##get median parameter estimates head(mcmc)
(Intercept) SEASONSpring SEASONSummer SEASONWinter DENSITYLow SEASONSpring:DENSITYLow 1 5.121259 -2.9272265 0.6091165 -3.139551 0.45113908 0.5706076 2 3.976797 -0.8830568 3.3102172 -2.330273 0.70468774 -0.1504981 3 4.115700 -1.9179433 2.5778731 -2.000068 0.47173325 0.3961598 4 3.533363 -1.1085150 3.3148439 -2.517882 1.04706721 -0.7040375 5 4.490729 -1.6438569 2.3067727 -1.947001 0.05754433 0.0959790 6 3.645614 -0.5453875 2.8895704 -1.325671 0.71087022 -0.4695005 SEASONSummer:DENSITYLow SEASONWinter:DENSITYLow sigma2 1 -2.364461 -1.0926231 1.5431413 2 -2.777595 -1.2870478 2.3160621 3 -2.333801 -1.8342519 1.0830885 4 -2.213357 -0.9719073 1.5486151 5 -2.377307 -0.6344449 1.1172891 6 -2.427330 -1.5042575 0.8387686
wch = grepl('sigma2',colnames(mcmc))==0 coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = quinn$SQRTRECRUITS - fit ggplot() + geom_point(quinn=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=SEASON))

ggplot(newdata) + geom_point(aes(y=resid, x=DENSITY))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model wch = grepl('sigma',colnames(mcmc))==0 coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~SEASON*DENSITY, data=quinn) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(quinn), fit[i,], sqrt(mcmc[i, 'sigma2']))) newdata = data.frame(SEASON=quinn$SEASON, DENSITY=quinn$DENSITY, yRep) %>% gather(key=Sample, value=Value,-SEASON,-DENSITY) ggplot(newdata) + geom_violin(aes(y=Value, x=SEASON, color=DENSITY,fill='Model'), alSEASONa=0.5)+ geom_violin(data=quinn, aes(y=SQRTRECRUITS,x=SEASON,color=DENSITY,fill='Obs'), alSEASONa=0.5) + geom_point(data=quinn, aes(y=SQRTRECRUITS, x=SEASON, color=DENSITY), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=DENSITY, fill='Model', group=DENSITY, color=SEASON), alSEASONa=0.5)+ geom_point(data=quinn, aes(y=SQRTRECRUITS, x=DENSITY, group=DENSITY,color=SEASON))

mcmc_intervals(as.matrix(quinn.mcmcpack), regex_pars='Intercept|^SEASON|^DENSITY|sigma')

mcmc_areas(as.matrix(quinn.mcmcpack), regex_pars='Intercept|^SEASON|^DENSITY|sigma')

mcmc = quinn.r2jags$BUGSoutput$sims.matrix #generate a model matrix newdata = quinn Xmat = model.matrix(~SEASON*DENSITY, newdata) ##get median parameter estimates head(mcmc)
beta[1] beta[2] beta[3] beta[4] beta[5] beta[6] beta[7] beta[8] deviance [1,] 3.650540 -1.5209859 3.084760 -1.398461 0.4047201 -0.05072334 -2.4858544 -0.7625953 124.5785 [2,] 4.289732 -1.1917061 3.247703 -1.960065 -0.2790781 -0.06967586 -3.0663729 -0.3547660 120.0191 [3,] 5.009456 -1.5737975 1.912638 -2.525833 -0.7566606 0.80796974 -1.6121361 -0.3474418 117.7514 [4,] 4.762701 -1.0146264 2.012612 -1.850269 -1.2065498 0.57783132 -0.4480735 -0.6276987 122.6503 [5,] 4.247299 -1.0707052 2.715825 -2.399065 -0.1095194 0.66114619 -1.5097850 -0.6635236 117.6268 [6,] 4.607514 -0.4451694 2.573628 -3.472830 -0.3669575 -0.92034494 -2.6014757 0.7713595 132.4770 sigma [1,] 1.0676190 [2,] 0.9208332 [3,] 1.0197379 [4,] 1.0568938 [5,] 1.0634309 [6,] 1.3488744wch = grep('^beta\\[',colnames(mcmc)) coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = quinn$SQRTRECRUITS - fit ggplot() + geom_point(quinn=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=SEASON))

ggplot(newdata) + geom_point(aes(y=resid, x=DENSITY))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~SEASON*DENSITY, data=quinn) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(quinn), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(SEASON=quinn$SEASON, DENSITY=quinn$DENSITY, yRep) %>% gather(key=Sample, value=Value,-SEASON,-DENSITY) ggplot(newdata) + geom_violin(aes(y=Value, x=SEASON, color=DENSITY,fill='Model'), alSEASONa=0.5)+ geom_violin(data=quinn, aes(y=SQRTRECRUITS,x=SEASON,color=DENSITY,fill='Obs'), alSEASONa=0.5) + geom_point(data=quinn, aes(y=SQRTRECRUITS, x=SEASON, color=DENSITY), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=DENSITY, fill='Model', group=DENSITY, color=SEASON), alSEASONa=0.5)+ geom_point(data=quinn, aes(y=SQRTRECRUITS, x=DENSITY, group=DENSITY,color=SEASON))

mcmc_intervals(as.matrix(quinn.mcmcpack), regex_pars='Intercept|^SEASON|^DENSITY|sigma')

mcmc_areas(as.matrix(quinn.mcmcpack), regex_pars='Intercept|^SEASON|^DENSITY|sigma')

mcmc = as.matrix(quinn.rstan) #generate a model matrix newdata = quinn Xmat = model.matrix(~SEASON*DENSITY, newdata) ##get median parameter estimates wch = grep('^beta\\[',colnames(mcmc)) coefs = apply(mcmc[,wch], 2, median) fit = as.vector(coefs %*% t(Xmat)) resid = quinn$SQRTRECRUITS - fit ggplot() + geom_point(quinn=NULL, aes(y=resid, x=fit))

newdata = newdata %>% cbind(fit,resid) ggplot(newdata) + geom_point(aes(y=resid, x=SEASON))

ggplot(newdata) + geom_point(aes(y=resid, x=DENSITY))

sresid = resid/sd(resid) ggplot() + geom_point(data=NULL, aes(y=sresid, x=fit))

## draw samples from this model coefs = as.matrix(mcmc[,wch]) Xmat = model.matrix(~SEASON*DENSITY, data=quinn) fit = coefs %*% t(Xmat) yRep = sapply(1:nrow(mcmc), function(i) rnorm(nrow(quinn), fit[i,], mcmc[i, 'sigma'])) newdata = data.frame(SEASON=quinn$SEASON, DENSITY=quinn$DENSITY, yRep) %>% gather(key=Sample, value=Value,-SEASON,-DENSITY) ggplot(newdata) + geom_violin(aes(y=Value, x=SEASON, color=DENSITY,fill='Model'), alSEASONa=0.5)+ geom_violin(data=quinn, aes(y=SQRTRECRUITS,x=SEASON,color=DENSITY,fill='Obs'), alSEASONa=0.5) + geom_point(data=quinn, aes(y=SQRTRECRUITS, x=SEASON, color=DENSITY), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=DENSITY, fill='Model', group=DENSITY, color=SEASON), alSEASONa=0.5)+ geom_point(data=quinn, aes(y=SQRTRECRUITS, x=DENSITY, group=DENSITY,color=SEASON))

mcmc_intervals(as.matrix(quinn.rstan), regex_pars='^beta|sigma')

mcmc_areas(as.matrix(quinn.rstan), regex_pars='^beta|sigma')

resid = resid(quinn.rstanarm) fit = fitted(quinn.rstanarm) ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

resid = resid(quinn.rstanarm) dat = quinn %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = SEASON))

ggplot(dat) + geom_point(aes(y = resid, x = DENSITY))

resid = resid(quinn.rstanarm) sigma(quinn.rstanarm)
[1] 1.031223
sresid = resid/sigma(quinn.rstanarm) fit = fitted(quinn.rstanarm) ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

y_pred = posterior_predict(quinn.rstanarm) newdata = quinn %>% dplyr:::select(-GROUP) %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -SEASON,-DENSITY,-SQRTRECRUITS) head(newdata)
SEASON DENSITY SQRTRECRUITS Rep Value 1 Spring Low 3.872983 RECRUITS 15 2 Spring Low 3.162278 RECRUITS 10 3 Spring Low 3.605551 RECRUITS 13 4 Spring Low 3.605551 RECRUITS 13 5 Spring Low 2.236068 RECRUITS 5 6 Spring High 3.316625 RECRUITS 11
ggplot(newdata) + geom_violin(aes(y=Value, x=SEASON, color=DENSITY,fill='Model'), alSEASONa=0.5)+ geom_violin(data=quinn, aes(y=SQRTRECRUITS,x=SEASON,color=DENSITY,fill='Obs'), alSEASONa=0.5) + geom_point(data=quinn, aes(y=SQRTRECRUITS, x=SEASON, color=DENSITY), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=DENSITY, fill='Model', group=DENSITY, color=SEASON), alSEASONa=0.5)+ geom_point(data=quinn, aes(y=SQRTRECRUITS, x=DENSITY, group=DENSITY,color=SEASON))

mcmc_intervals(as.matrix(quinn.rstanarm), regex_pars='Intercept|^SEASON|^DENSITY|sigma')

mcmc_areas(as.matrix(quinn.rstanarm), regex_pars='Intercept|^SEASON|^DENSITY|sigma')
 All diagnostics seem reasonable.
All diagnostics seem reasonable.resid = resid(quinn.brms)[,'Estimate'] fit = fitted(quinn.brms)[,'Estimate'] ggplot() + geom_point(data = NULL, aes(y = resid, x = fit))

resid = resid(quinn.brms)[,'Estimate'] dat = quinn %>% mutate(resid = resid) ggplot(dat) + geom_point(aes(y = resid, x = SEASON))

ggplot(dat) + geom_point(aes(y = resid, x = DENSITY))

resid = resid(quinn.brms) sresid = resid(quinn.brms, type='pearson')[,'Estimate'] fit = fitted(quinn.brms)[,'Estimate'] ggplot() + geom_point(data = NULL, aes(y = sresid, x = fit))

y_pred = posterior_predict(quinn.brms) newdata = quinn %>% dplyr:::select(-GROUP) %>% cbind(t(y_pred)) %>% gather(key = "Rep", value = "Value", -SEASON,-DENSITY,-SQRTRECRUITS) head(newdata)
SEASON DENSITY SQRTRECRUITS Rep Value 1 Spring Low 3.872983 RECRUITS 15 2 Spring Low 3.162278 RECRUITS 10 3 Spring Low 3.605551 RECRUITS 13 4 Spring Low 3.605551 RECRUITS 13 5 Spring Low 2.236068 RECRUITS 5 6 Spring High 3.316625 RECRUITS 11
ggplot(newdata) + geom_violin(aes(y=Value, x=SEASON, color=DENSITY,fill='Model'), alSEASONa=0.5)+ geom_violin(data=quinn, aes(y=SQRTRECRUITS,x=SEASON,color=DENSITY,fill='Obs'), alSEASONa=0.5) + geom_point(data=quinn, aes(y=SQRTRECRUITS, x=SEASON, color=DENSITY), position=position_jitter(width=0.1,height=0))

ggplot(newdata) + geom_violin(aes(y=Value, x=DENSITY, fill='Model', group=DENSITY, color=SEASON), alSEASONa=0.5)+ geom_point(data=quinn, aes(y=SQRTRECRUITS, x=DENSITY, group=DENSITY,color=SEASON))

mcmc_intervals(as.matrix(quinn.brms), regex_pars='^b_|sigma')

mcmc_areas(as.matrix(quinn.brms), regex_pars='^b_|sigma')

- Explore parameter estimates
summary(quinn.mcmcpack)
Iterations = 1001:11000 Thinning interval = 1 Number of chains = 1 Sample size per chain = 10000 1. Empirical mean and standard deviation for each variable, plus standard error of the mean: Mean SD Naive SE Time-series SE (Intercept) 4.22521 0.4242 0.004242 0.004242 SEASONSpring -1.20962 0.6065 0.006065 0.005909 SEASONSummer 2.64176 0.6046 0.006046 0.006046 SEASONWinter -1.95379 0.5951 0.005951 0.005951 DENSITYLow 0.03276 0.6641 0.006641 0.006641 SEASONSpring:DENSITYLow 0.24480 0.9229 0.009229 0.008858 SEASONSummer:DENSITYLow -2.25004 0.8997 0.008997 0.008997 SEASONWinter:DENSITYLow -1.35434 0.9852 0.009852 0.009602 sigma2 1.07937 0.2771 0.002771 0.003376 2. Quantiles for each variable: 2.5% 25% 50% 75% 97.5% (Intercept) 3.4057 3.9434 4.22488 4.4979 5.074057 SEASONSpring -2.4195 -1.6076 -1.21069 -0.8090 -0.001834 SEASONSummer 1.4456 2.2436 2.64736 3.0391 3.826575 SEASONWinter -3.1102 -2.3412 -1.96053 -1.5590 -0.774191 DENSITYLow -1.2961 -0.4019 0.03212 0.4669 1.343547 SEASONSpring:DENSITYLow -1.5708 -0.3584 0.24801 0.8475 2.068431 SEASONSummer:DENSITYLow -4.0197 -2.8466 -2.24992 -1.6559 -0.497425 SEASONWinter:DENSITYLow -3.2916 -2.0105 -1.34501 -0.6982 0.554175 sigma2 0.6726 0.8807 1.03389 1.2285 1.739985#OR library(broom) tidyMCMC(quinn.mcmcpack,conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 (Intercept) 4.22521285 0.4241621 3.3943276 5.05487056 2 SEASONSpring -1.20962463 0.6065284 -2.4480233 -0.04081777 3 SEASONSummer 2.64175924 0.6045629 1.4442037 3.82327523 4 SEASONWinter -1.95378953 0.5951092 -3.1219475 -0.78794486 5 DENSITYLow 0.03276479 0.6641271 -1.3328321 1.27378209 6 SEASONSpring:DENSITYLow 0.24479898 0.9229198 -1.5970338 2.03223172 7 SEASONSummer:DENSITYLow -2.25004162 0.8996546 -4.0356145 -0.51881764 8 SEASONWinter:DENSITYLow -1.35434133 0.9852031 -3.2702655 0.56471620 9 sigma2 1.07936870 0.2771246 0.6299837 1.64193631
#OR with p-values newdata = with(quinn, expand.grid(SEASON=levels(SEASON), DENSITY=levels(DENSITY))) Xmat = model.matrix(~SEASON*DENSITY, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(quinn.mcmcpack[,i]) )
[1] 0 [1] 0.0498 [1] 1e-04 [1] 0.0018 [1] 0.9573 [1] 0.7864 [1] 0.0142 [1] 0.1644
# Main effect of SEASON mcmcpvalue(quinn.mcmcpack[,which(wch==1)])
[1] 0
# Main effect of DENSITY mcmcpvalue(quinn.mcmcpack[,which(wch==2)])
[1] 0.9573
# Interaction mcmcpvalue(quinn.mcmcpack[,which(wch==3)])
[1] 0.0206
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08anova(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) SEASON 3 87.454 29.1512 28.5740 2.067e-09 *** DENSITY 1 7.089 7.0894 6.9491 0.01255 * SEASON:DENSITY 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(SQRTRECRUITS~DENSITY*SEASON, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) DENSITY 1 2.854 2.8541 2.7976 0.10359 SEASON 3 91.689 30.5630 29.9579 1.165e-09 *** DENSITY:SEASON 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1print(quinn.r2jags)
Inference for Bugs model at "5", fit using jags, 3 chains, each with 50000 iterations (first 3000 discarded), n.thin = 10 n.sims = 14100 iterations saved mu.vect sd.vect 2.5% 25% 50% 75% 97.5% Rhat n.eff beta[1] 4.228 0.430 3.380 3.938 4.232 4.518 5.059 1.001 14000 beta[2] -1.203 0.606 -2.380 -1.608 -1.203 -0.805 -0.004 1.001 14000 beta[3] 2.646 0.611 1.467 2.244 2.639 3.041 3.853 1.001 14000 beta[4] -1.954 0.609 -3.120 -2.364 -1.953 -1.550 -0.756 1.001 14000 beta[5] 0.038 0.686 -1.308 -0.419 0.037 0.497 1.380 1.001 7700 beta[6] 0.233 0.941 -1.613 -0.391 0.223 0.865 2.069 1.001 14000 beta[7] -2.257 0.923 -4.066 -2.866 -2.265 -1.646 -0.446 1.001 9200 beta[8] -1.366 1.009 -3.366 -2.035 -1.363 -0.702 0.601 1.001 4800 sigma 1.051 0.135 0.826 0.955 1.036 1.131 1.355 1.001 14000 deviance 121.649 4.950 114.364 118.027 120.859 124.483 133.285 1.001 11000 For each parameter, n.eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor (at convergence, Rhat=1). DIC info (using the rule, pD = var(deviance)/2) pD = 12.3 and DIC = 133.9 DIC is an estimate of expected predictive error (lower deviance is better).#OR library(broom) tidyMCMC(quinn.r2jags$BUGSoutput$sims.matrix,conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 beta[1] 4.22750954 0.4300189 3.3871678 5.062626384 2 beta[2] -1.20342419 0.6055745 -2.3664669 0.007509219 3 beta[3] 2.64612198 0.6106636 1.4496400 3.834212836 4 beta[4] -1.95393888 0.6086991 -3.1674486 -0.809400274 5 beta[5] 0.03784565 0.6864609 -1.2666616 1.414769981 6 beta[6] 0.23343378 0.9405136 -1.5551796 2.117044828 7 beta[7] -2.25699115 0.9226389 -4.0689985 -0.452625591 8 beta[8] -1.36584749 1.0089406 -3.3079949 0.654825737 9 deviance 121.64897632 4.9499792 113.4283894 131.360192312 10 sigma 1.05077704 0.1354712 0.8097102 1.327079784
#OR with p-values newdata = with(quinn, expand.grid(SEASON=levels(SEASON), DENSITY=levels(DENSITY))) Xmat = model.matrix(~SEASON*DENSITY, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(quinn.r2jags$BUGSoutput$sims.matrix[,i]) )
[1] 0 [1] 0.04794326 [1] 0.0002836879 [1] 0.002836879 [1] 0.9568794 [1] 0.8021986 [1] 0.01624113 [1] 0.170922
# Main effect of SEASON mcmcpvalue(quinn.r2jags$BUGSoutput$sims.matrix[,which(wch==1)])
[1] 0
# Main effect of DENSITY mcmcpvalue(quinn.r2jags$BUGSoutput$sims.matrix[,which(wch==2)])
[1] 0.9568794
# Interaction mcmcpvalue(quinn.r2jags$BUGSoutput$sims.matrix[,which(wch==3)])
[1] 0.02531915
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08anova(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) SEASON 3 87.454 29.1512 28.5740 2.067e-09 *** DENSITY 1 7.089 7.0894 6.9491 0.01255 * SEASON:DENSITY 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(SQRTRECRUITS~DENSITY*SEASON, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) DENSITY 1 2.854 2.8541 2.7976 0.10359 SEASON 3 91.689 30.5630 29.9579 1.165e-09 *** DENSITY:SEASON 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1print(quinn.rstan, pars=c('beta','sigma'))
Inference for Stan model: a6ade6a40138b983c8773faeb7752a0c. 3 chains, each with iter=2000; warmup=500; thin=3; post-warmup draws per chain=500, total post-warmup draws=1500. mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat beta[1] 4.25 0.01 0.43 3.37 3.97 4.25 4.53 5.05 1122 1 beta[2] -1.24 0.02 0.59 -2.44 -1.63 -1.22 -0.83 -0.15 1023 1 beta[3] 2.61 0.02 0.60 1.38 2.21 2.60 3.01 3.86 788 1 beta[4] -1.98 0.02 0.61 -3.16 -2.38 -1.99 -1.57 -0.76 1062 1 beta[5] -0.01 0.02 0.66 -1.30 -0.45 -0.03 0.42 1.30 1007 1 beta[6] 0.29 0.03 0.92 -1.50 -0.30 0.28 0.90 2.10 1067 1 beta[7] -2.19 0.03 0.90 -3.93 -2.77 -2.19 -1.59 -0.45 1107 1 beta[8] -1.31 0.03 0.97 -3.19 -1.95 -1.33 -0.64 0.56 1270 1 sigma 1.04 0.00 0.13 0.83 0.95 1.03 1.11 1.32 1473 1 Samples were drawn using NUTS(diag_e) at Tue Dec 19 08:18:10 2017. For each parameter, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat=1).#OR library(broom) tidyMCMC(quinn.rstan,conf.int=TRUE, conf.method='HPDinterval', pars=c('beta','sigma'))
term estimate std.error conf.low conf.high 1 beta[1] 4.248943796 0.4254124 3.4227812 5.0804326 2 beta[2] -1.241198464 0.5934883 -2.3956122 -0.1251383 3 beta[3] 2.607283964 0.5994328 1.3341528 3.7755285 4 beta[4] -1.980857954 0.6065078 -3.1675146 -0.7992811 5 beta[5] -0.007740026 0.6620968 -1.4105816 1.1773974 6 beta[6] 0.292309186 0.9171744 -1.4589296 2.1106305 7 beta[7] -2.193772814 0.8969077 -3.8916079 -0.4352106 8 beta[8] -1.313050558 0.9735668 -3.2755647 0.4342699 9 sigma 1.039876545 0.1275896 0.8146716 1.2908550
#OR with p-values newdata = with(quinn, expand.grid(SEASON=levels(SEASON), DENSITY=levels(DENSITY))) Xmat = model.matrix(~SEASON*DENSITY, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(quinn.rstan)[,i]) )
[1] 0 [1] 0.03866667 [1] 0 [1] 0 [1] 0.992 [1] 0.736 [1] 0.02333333 [1] 0.178
# Main effect of SEASON mcmcpvalue(as.matrix(quinn.rstan)[,which(wch==1)])
[1] 0
# Main effect of DENSITY mcmcpvalue(as.matrix(quinn.rstan)[,which(wch==2)])
[1] 0.992
# Interaction mcmcpvalue(as.matrix(quinn.rstan)[,which(wch==3)])
[1] 0.02266667
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08anova(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) SEASON 3 87.454 29.1512 28.5740 2.067e-09 *** DENSITY 1 7.089 7.0894 6.9491 0.01255 * SEASON:DENSITY 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(SQRTRECRUITS~DENSITY*SEASON, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) DENSITY 1 2.854 2.8541 2.7976 0.10359 SEASON 3 91.689 30.5630 29.9579 1.165e-09 *** DENSITY:SEASON 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(extract_log_lik(quinn.rstan)))
Computed from 1500 by 42 log-likelihood matrix Estimate SE elpd_loo -66.3 5.0 p_loo 8.9 2.1 looic 132.5 10.1 Pareto k diagnostic values: Count Pct (-Inf, 0.5] (good) 36 85.7% (0.5, 0.7] (ok) 5 11.9% (0.7, 1] (bad) 1 2.4% (1, Inf) (very bad) 0 0.0% See help('pareto-k-diagnostic') for details.# now fit a model without main factor modelString=" data { int<lower=1> n; int<lower=1> nX; vector [n] y; matrix [n,nX] X; } parameters { vector[nX] beta; real<lower=0> sigma; } transformed parameters { vector[n] mu; mu = X*beta; } model { #Likelihood y~normal(mu,sigma); #Priors beta ~ normal(0,1000); sigma~cauchy(0,5); } generated quantities { vector[n] log_lik; for (i in 1:n) { log_lik[i] = normal_lpdf(y[i] | mu[i], sigma); } } " Xmat <- model.matrix(~SEASON+DENSITY, quinn) quinn.list <- with(quinn,list(y=SQRTRECRUITS, X=Xmat,n=nrow(quinn), nX=ncol(Xmat))) quinn.rstan.red <- stan(data=quinn.list, model_code=modelString, chains=3, iter=2000, warmup=500, thin=3, refresh=FALSE )
In file included from /usr/local/lib/R/site-library/BH/include/boost/config.hpp:39:0, from /usr/local/lib/R/site-library/BH/include/boost/math/tools/config.hpp:13, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/var.hpp:7, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core/gevv_vvv_vari.hpp:5, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/core.hpp:12, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math/rev/mat.hpp:4, from /usr/local/lib/R/site-library/StanHeaders/include/stan/math.hpp:4, from /usr/local/lib/R/site-library/StanHeaders/include/src/stan/model/model_header.hpp:4, from file79605948c4cc.cpp:8: /usr/local/lib/R/site-library/BH/include/boost/config/compiler/gcc.hpp:186:0: warning: "BOOST_NO_CXX11_RVALUE_REFERENCES" redefined # define BOOST_NO_CXX11_RVALUE_REFERENCES ^ <command-line>:0:0: note: this is the location of the previous definition Gradient evaluation took 1.7e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.17 seconds. Adjust your expectations accordingly! Elapsed Time: 0.028322 seconds (Warm-up) 0.07944 seconds (Sampling) 0.107762 seconds (Total) Gradient evaluation took 8e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.08 seconds. Adjust your expectations accordingly! Elapsed Time: 0.027095 seconds (Warm-up) 0.070301 seconds (Sampling) 0.097396 seconds (Total) Gradient evaluation took 8e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.08 seconds. Adjust your expectations accordingly! Elapsed Time: 0.027192 seconds (Warm-up) 0.066441 seconds (Sampling) 0.093633 seconds (Total)(reduced=loo(extract_log_lik(quinn.rstan.red)))
Computed from 1500 by 42 log-likelihood matrix Estimate SE elpd_loo -68.0 4.7 p_loo 5.6 1.3 looic 136.1 9.3 Pareto k diagnostic values: Count Pct (-Inf, 0.5] (good) 41 97.6% (0.5, 0.7] (ok) 1 2.4% (0.7, 1] (bad) 0 0.0% (1, Inf) (very bad) 0 0.0% All Pareto k estimates are ok (k < 0.7) See help('pareto-k-diagnostic') for details.par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
summary(quinn.rstanarm)
Model Info: function: stan_glm family: gaussian [identity] formula: SQRTRECRUITS ~ SEASON * DENSITY algorithm: sampling priors: see help('prior_summary') sample: 2250 (posterior sample size) num obs: 42 Estimates: mean sd 2.5% 25% 50% 75% 97.5% (Intercept) 4.3 0.4 3.6 4.0 4.3 4.6 5.1 SEASONSpring -1.2 0.5 -2.2 -1.6 -1.2 -0.8 -0.1 SEASONSummer 2.4 0.5 1.3 2.0 2.4 2.7 3.4 SEASONWinter -2.0 0.5 -3.0 -2.3 -2.0 -1.6 -0.9 DENSITYLow -0.2 0.5 -1.2 -0.6 -0.2 0.1 0.9 SEASONSpring:DENSITYLow 0.4 0.8 -1.2 -0.1 0.4 0.9 1.8 SEASONSummer:DENSITYLow -1.7 0.8 -3.2 -2.2 -1.7 -1.2 -0.2 SEASONWinter:DENSITYLow -1.1 0.8 -2.7 -1.6 -1.0 -0.5 0.5 sigma 1.0 0.1 0.8 0.9 1.0 1.1 1.3 mean_PPD 3.9 0.2 3.4 3.7 3.9 4.0 4.3 log-posterior -76.1 2.4 -81.7 -77.4 -75.7 -74.3 -72.5 Diagnostics: mcse Rhat n_eff (Intercept) 0.0 1.0 1679 SEASONSpring 0.0 1.0 1762 SEASONSummer 0.0 1.0 1737 SEASONWinter 0.0 1.0 1930 DENSITYLow 0.0 1.0 1563 SEASONSpring:DENSITYLow 0.0 1.0 1735 SEASONSummer:DENSITYLow 0.0 1.0 1501 SEASONWinter:DENSITYLow 0.0 1.0 1872 sigma 0.0 1.0 1945 mean_PPD 0.0 1.0 2160 log-posterior 0.1 1.0 1467 For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).#OR library(broom) tidyMCMC(quinn.rstanarm$stanfit,conf.int=TRUE, conf.method='HPDinterval', rhat=TRUE, ess=TRUE)
term estimate std.error conf.low conf.high rhat ess 1 (Intercept) 4.2944621 0.3781257 3.6008854 5.0780981 1.0001016 1679 2 SEASONSpring -1.1944391 0.5415104 -2.2425511 -0.1488528 1.0007214 1762 3 SEASONSummer 2.3506163 0.5418177 1.2919129 3.4111662 1.0019149 1737 4 SEASONWinter -1.9593821 0.5441314 -3.0178154 -0.8888779 1.0000743 1930 5 DENSITYLow -0.2031083 0.5393479 -1.2823813 0.8410013 1.0000778 1563 6 SEASONSpring:DENSITYLow 0.3687119 0.7663820 -1.1687244 1.8007469 1.0001554 1735 7 SEASONSummer:DENSITYLow -1.6934501 0.7516600 -3.2933392 -0.2561633 1.0005900 1501 8 SEASONWinter:DENSITYLow -1.0512296 0.8304569 -2.6875590 0.4954263 0.9990874 1872 9 sigma 1.0430770 0.1315636 0.8032712 1.2960622 0.9995744 1945 10 mean_PPD 3.8718671 0.2251886 3.4482842 4.3225541 1.0000943 2160 11 log-posterior -76.0590659 2.3820147 -80.9909582 -72.2228945 0.9989467 1467
#OR with p-values newdata = with(quinn, expand.grid(SEASON=levels(SEASON), DENSITY=levels(DENSITY))) Xmat = model.matrix(~SEASON*DENSITY, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(quinn.rstanarm)[,i]) )
[1] 0 [1] 0.03066667 [1] 0 [1] 0.0004444444 [1] 0.6968889 [1] 0.636 [1] 0.02977778 [1] 0.2071111
# Main effect of SEASON mcmcpvalue(as.matrix(quinn.rstanarm)[,which(wch==1)])
[1] 0
# Main effect of DENSITY mcmcpvalue(as.matrix(quinn.rstanarm)[,which(wch==2)])
[1] 0.6968889
# Interaction mcmcpvalue(as.matrix(quinn.rstanarm)[,which(wch==3)])
[1] 0.05155556
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08anova(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) SEASON 3 87.454 29.1512 28.5740 2.067e-09 *** DENSITY 1 7.089 7.0894 6.9491 0.01255 * SEASON:DENSITY 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(SQRTRECRUITS~DENSITY*SEASON, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) DENSITY 1 2.854 2.8541 2.7976 0.10359 SEASON 3 91.689 30.5630 29.9579 1.165e-09 *** DENSITY:SEASON 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(quinn.rstanarm))
Computed from 2250 by 42 log-likelihood matrix Estimate SE elpd_loo -65.5 4.8 p_loo 8.0 1.7 looic 131.1 9.6 Pareto k diagnostic values: Count Pct (-Inf, 0.5] (good) 36 85.7% (0.5, 0.7] (ok) 6 14.3% (0.7, 1] (bad) 0 0.0% (1, Inf) (very bad) 0 0.0% All Pareto k estimates are ok (k < 0.7) See help('pareto-k-diagnostic') for details.quinn.rstanarm.red = update(quinn.rstanarm, .~SEASON+DENSITY)
Gradient evaluation took 5.5e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.55 seconds. Adjust your expectations accordingly! Elapsed Time: 0.033283 seconds (Warm-up) 0.08671 seconds (Sampling) 0.119993 seconds (Total) Gradient evaluation took 1.3e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.13 seconds. Adjust your expectations accordingly! Elapsed Time: 0.03061 seconds (Warm-up) 0.092371 seconds (Sampling) 0.122981 seconds (Total) Gradient evaluation took 1.4e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.14 seconds. Adjust your expectations accordingly! Elapsed Time: 0.036775 seconds (Warm-up) 0.078319 seconds (Sampling) 0.115094 seconds (Total)(reduced=loo(quinn.rstanarm.red))
Computed from 2250 by 42 log-likelihood matrix Estimate SE elpd_loo -67.9 4.6 p_loo 5.5 1.2 looic 135.8 9.2 All Pareto k estimates are good (k < 0.5) See help('pareto-k-diagnostic') for details.par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
elpd_diff se -2.4 2.4There is very some support for an interaction (loo of the full model is less than that of the reduced model). Interaction effects suggest that recruitment is higher in the Summer low density population than would be expected if there was no interactions.summary(quinn.brms)
Family: gaussian(identity) Formula: SQRTRECRUITS ~ SEASON * DENSITY Data: quinn (Number of observations: 42) Samples: 3 chains, each with iter = 2000; warmup = 500; thin = 2; total post-warmup samples = 2250 ICs: LOO = NA; WAIC = NA; R2 = NA Population-Level Effects: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat Intercept 4.29 0.33 3.63 4.93 1854 1 SEASONSpring -1.02 0.46 -1.91 -0.07 1884 1 SEASONSummer 2.00 0.48 1.02 2.92 1887 1 SEASONWinter -1.83 0.46 -2.75 -0.95 1941 1 DENSITYLow -0.37 0.42 -1.19 0.41 1890 1 SEASONSpring:DENSITYLow 0.31 0.60 -0.87 1.50 1962 1 SEASONSummer:DENSITYLow -1.07 0.60 -2.19 0.15 1979 1 SEASONWinter:DENSITYLow -0.85 0.65 -2.15 0.34 1939 1 Family Specific Parameters: Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat sigma 1.06 0.14 0.83 1.36 1776 1 Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence, Rhat = 1).#OR library(broom) tidyMCMC(quinn.brms$fit,conf.int=TRUE, conf.method='HPDinterval', rhat=TRUE, ess=TRUE)
term estimate std.error conf.low conf.high rhat ess 1 b_Intercept 4.2937368 0.3267812 3.6208843 4.92899297 1.0009963 1854 2 b_SEASONSpring -1.0204416 0.4627238 -1.8869003 -0.05501539 1.0003437 1884 3 b_SEASONSummer 1.9975818 0.4765084 1.1050091 2.99362386 0.9989374 1887 4 b_SEASONWinter -1.8325778 0.4591678 -2.6768481 -0.88445027 1.0004183 1941 5 b_DENSITYLow -0.3696387 0.4230952 -1.2161672 0.38096496 1.0016643 1890 6 b_SEASONSpring:DENSITYLow 0.3098758 0.5999900 -0.9051438 1.44819946 1.0001602 1962 7 b_SEASONSummer:DENSITYLow -1.0658827 0.6002369 -2.2027007 0.12982398 0.9991428 1979 8 b_SEASONWinter:DENSITYLow -0.8468540 0.6453871 -2.1293147 0.35870271 0.9990898 1939 9 sigma 1.0602483 0.1361341 0.8138045 1.33499319 1.0003507 1776
#OR with p-values newdata = with(quinn, expand.grid(SEASON=levels(SEASON), DENSITY=levels(DENSITY))) Xmat = model.matrix(~SEASON*DENSITY, data=newdata) wch = attr(Xmat, 'assign') for (i in 1:ncol(Xmat)) print(mcmcpvalue(as.matrix(quinn.brms)[,i]) )
[1] 0 [1] 0.03066667 [1] 0 [1] 0.0004444444 [1] 0.3906667 [1] 0.5951111 [1] 0.07377778 [1] 0.1871111
# Main effect of SEASON mcmcpvalue(as.matrix(quinn.brms)[,which(wch==1)])
[1] 0
# Main effect of DENSITY mcmcpvalue(as.matrix(quinn.brms)[,which(wch==2)])
[1] 0.3906667
# Interaction mcmcpvalue(as.matrix(quinn.brms)[,which(wch==3)])
[1] 0.1582222
## frequentist for comparison - notice the issue ## due to imbalance and Type I SS summary(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08anova(lm(SQRTRECRUITS~SEASON*DENSITY, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) SEASON 3 87.454 29.1512 28.5740 2.067e-09 *** DENSITY 1 7.089 7.0894 6.9491 0.01255 * SEASON:DENSITY 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1anova(lm(SQRTRECRUITS~DENSITY*SEASON, data=quinn))
Analysis of Variance Table Response: SQRTRECRUITS Df Sum Sq Mean Sq F value Pr(>F) DENSITY 1 2.854 2.8541 2.7976 0.10359 SEASON 3 91.689 30.5630 29.9579 1.165e-09 *** DENSITY:SEASON 3 11.354 3.7848 3.7098 0.02068 * Residuals 34 34.687 1.0202 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Compare loo library(loo) (full=loo(quinn.brms))
LOOIC SE 131.34 9.54
quinn.brms.red = update(quinn.brms, .~SEASON+DENSITY, refresh=FALSE)
Gradient evaluation took 1.6e-05 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.16 seconds. Adjust your expectations accordingly! Elapsed Time: 0.036558 seconds (Warm-up) 0.033247 seconds (Sampling) 0.069805 seconds (Total) Gradient evaluation took 9e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds. Adjust your expectations accordingly! Elapsed Time: 0.039014 seconds (Warm-up) 0.031361 seconds (Sampling) 0.070375 seconds (Total) Gradient evaluation took 9e-06 seconds 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds. Adjust your expectations accordingly! Elapsed Time: 0.038403 seconds (Warm-up) 0.030969 seconds (Sampling) 0.069372 seconds (Total)(reduced=loo(quinn.brms.red))
LOOIC SE 135.56 9.08
par(mfrow = 1:2, mar = c(5,3.8,1,0) + 0.1, las = 3) plot(full, label_points = TRUE) plot(reduced, label_points = TRUE)

compare_models(full,reduced)
Error in discrete == discrete[1]: comparison of these types is not implemented
- Explore the general effect of DENSITY separately in each of the SEASONs. We will start by exploring effect
sizes in sqrt units and percent differences between high and low density. We will also explore backtransformed effect sizes.
Note, backtransformations in the presence of effects that range from less than 0 to greater than 0, or less than 1 to greater than
one are not ideal as exponential is not a monotomic function across the entire number range.
mcmc = quinn.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^SEASON|^DENSITY", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) # Split the Xmat up according to SEASON # Subtract high density from low (since high comes # before low in the alphabet, high will be item 1 # bind into a single matrix Xmat = do.call('rbind',lapply(split(1:nrow(Xmat), newdata$SEASON),function(i) Xmat[i[1],] - Xmat[i[2],])) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn -0.03276479 0.6641271 -1.27378209 1.332832 2 Spring -0.27756376 0.6387045 -1.51996485 0.962308 3 Summer 2.21727684 0.5989693 1.01002298 3.359681 4 Winter 1.32157654 0.7360262 -0.09583538 2.781650
# OR if we express this as a percentage change Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) 100*(fit[i[1],] - fit[i[2],])/fit[i[2],]))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 0.7463853 16.33929 -30.27703 33.35491 2 Spring -6.4278375 19.81106 -41.04496 34.46112 3 Summer 48.9729376 16.79422 16.94628 81.92190 4 Winter 878.9675697 55887.13701 -1260.50116 2094.39264
tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval', estimate.method='median')
term estimate std.error conf.low conf.high 1 Autumn -0.7519079 16.33929 -30.27703 33.35491 2 Spring -8.3703722 19.81106 -41.04496 34.46112 3 Summer 47.5490983 16.79422 16.94628 81.92190 4 Winter 117.5129480 55887.13701 -1260.50116 2094.39264
## Express in natural units (rather than sqrt) Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) exp(fit[i[1],] - fit[i[2],])))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 1.209217 0.9237754 0.1186880 2.937502 2 Spring 0.930308 0.6858861 0.1329298 2.190961 3 Summer 10.998513 7.3966430 1.5499514 24.775873 4 Winter 4.920579 4.2974765 0.3034355 12.538327
mcmc = quinn.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) # Split the Xmat up according to SEASON # Subtract high density from low (since high comes # before low in the alphabet, high will be item 1 # bind into a single matrix Xmat = do.call('rbind',lapply(split(1:nrow(Xmat), newdata$SEASON),function(i) Xmat[i[1],] - Xmat[i[2],])) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn -0.03784565 0.6864609 -1.4147700 1.266662 2 Spring -0.27127943 0.6454227 -1.5326258 1.001519 3 Summer 2.21914550 0.6154643 1.0440081 3.472214 4 Winter 1.32800184 0.7436444 -0.1117116 2.815277
# OR if we express this as a percentage change Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) 100*(fit[i[1],] - fit[i[2],])/fit[i[2],]))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 0.7452126 16.90167 -30.74629 33.61524 2 Spring -6.1391619 20.10808 -42.56484 34.32284 3 Summer 49.0095454 17.20531 16.72296 83.74852 4 Winter 220.4362931 9659.02840 -1414.48216 1776.20659
tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval', estimate.method='median')
term estimate std.error conf.low conf.high 1 Autumn -0.8575932 16.90167 -30.74629 33.61524 2 Spring -8.3623325 20.10808 -42.56484 34.32284 3 Summer 47.7933806 17.20531 16.72296 83.74852 4 Winter 117.6845638 9659.02840 -1414.48216 1776.20659
## Express in natural units (rather than sqrt) Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) exp(fit[i[1],] - fit[i[2],])))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 1.2204372 0.9754432 0.1172933 2.995797 2 Spring 0.9407171 0.6918078 0.1096614 2.228955 3 Summer 11.1376557 7.9426863 1.5092637 25.461891 4 Winter 4.9899479 4.4003055 0.3220061 12.976154
mcmc = as.matrix(quinn.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) # Split the Xmat up according to SEASON # Subtract high density from low (since high comes # before low in the alphabet, high will be item 1 # bind into a single matrix Xmat = do.call('rbind',lapply(split(1:nrow(Xmat), newdata$SEASON),function(i) Xmat[i[1],] - Xmat[i[2],])) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 0.007740026 0.6620968 -1.1773974 1.410582 2 Spring -0.284569160 0.6359350 -1.4822238 1.001825 3 Summer 2.201512840 0.5973101 0.9684529 3.356481 4 Winter 1.320790584 0.7513553 -0.2961845 2.710948
# OR if we express this as a percentage change Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) 100*(fit[i[1],] - fit[i[2],])/fit[i[2],]))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 1.701494 16.31091 -27.68559 35.62625 2 Spring -6.708657 19.44638 -41.11105 33.04107 3 Summer 48.565539 16.67208 18.41655 83.67920 4 Winter -129.308975 12547.80716 -1638.76784 2050.07493
tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval', estimate.method='median')
term estimate std.error conf.low conf.high 1 Autumn 0.7840606 16.31091 -27.68559 35.62625 2 Spring -8.9801032 19.44638 -41.11105 33.04107 3 Summer 46.9827976 16.67208 18.41655 83.67920 4 Winter 117.9803984 12547.80716 -1638.76784 2050.07493
## Express in natural units (rather than sqrt) Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) exp(fit[i[1],] - fit[i[2],])))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 1.252313 0.9295281 0.1199441 3.037197 2 Spring 0.918678 0.6266919 0.1428231 2.171858 3 Summer 10.820446 7.2981230 1.4111153 24.039717 4 Winter 4.969052 4.4164902 0.3893657 13.279478
mcmc = as.matrix(quinn.rstanarm) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SEASON|^DENSITY',colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) # Split the Xmat up according to SEASON # Subtract high density from low (since high comes # before low in the alphabet, high will be item 1 # bind into a single matrix Xmat = do.call('rbind',lapply(split(1:nrow(Xmat), newdata$SEASON),function(i) Xmat[i[1],] - Xmat[i[2],])) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 0.2031083 0.5393479 -0.8410013 1.282381 2 Spring -0.1656036 0.6168246 -1.3472168 1.016781 3 Summer 1.8965584 0.5777641 0.7999547 3.105397 4 Winter 1.2543379 0.6843055 -0.1727148 2.511181
# OR if we express this as a percentage change Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) 100*(fit[i[1],] - fit[i[2],])/fit[i[2],]))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 6.172205 14.07126 -20.59639 35.48852 2 Spring -2.999168 19.86605 -37.10152 36.68237 3 Summer 41.033794 15.07603 10.38299 69.42044 4 Winter 58.014439 3503.90890 -54.99497 1703.18432
tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval', estimate.method='median')
term estimate std.error conf.low conf.high 1 Autumn 5.010770 14.07126 -20.59639 35.48852 2 Spring -5.305217 19.86605 -37.10152 36.68237 3 Summer 40.007645 15.07603 10.38299 69.42044 4 Winter 107.514361 3503.90890 -54.99497 1703.18432
## Express in natural units (rather than sqrt) Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) exp(fit[i[1],] - fit[i[2],])))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 1.412829 0.7933362 0.2380957 2.966709 2 Spring 1.028162 0.7192295 0.1612914 2.369805 3 Summer 7.843505 4.7466304 1.2035485 16.931993 4 Winter 4.411024 3.2739499 0.4755671 10.577953
mcmc = as.matrix(quinn.brms) wch = grep('^b_',colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) # Split the Xmat up according to SEASON # Subtract high density from low (since high comes # before low in the alphabet, high will be item 1 # bind into a single matrix Xmat = do.call('rbind',lapply(split(1:nrow(Xmat), newdata$SEASON),function(i) Xmat[i[1],] - Xmat[i[2],])) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 0.36963871 0.4230952 -0.38096496 1.216167 2 Spring 0.05976289 0.5564328 -1.10853575 1.082006 3 Summer 1.43552140 0.5421303 0.35218755 2.489011 4 Winter 1.21649267 0.6094468 0.02689828 2.455690
# OR if we express this as a percentage change Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) 100*(fit[i[1],] - fit[i[2],])/fit[i[2],]))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 10.333538 11.98208 -11.057890 34.00337 2 Spring 3.713672 18.56319 -30.086745 41.04957 3 Summer 30.431547 13.19500 4.422852 56.63755 4 Winter 155.963143 1360.16753 -25.640929 610.27027
tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval', estimate.method='median')
term estimate std.error conf.low conf.high 1 Autumn 9.254491 11.98208 -11.057890 34.00337 2 Spring 2.020509 18.56319 -30.086745 41.04957 3 Summer 29.605558 13.19500 4.422852 56.63755 4 Winter 94.980463 1360.16753 -25.640929 610.27027
## Express in natural units (rather than sqrt) Xmat = model.matrix(~SEASON*DENSITY,newdata) fit=coefs %*% t(Xmat) fit = t(fit) fit = t(do.call('rbind',lapply(split(1:nrow(fit), newdata$SEASON),function(i) exp(fit[i[1],] - fit[i[2],])))) tidyMCMC(as.mcmc(fit), conf.int=TRUE, conf.method='HPDinterval')
term estimate std.error conf.low conf.high 1 Autumn 1.585272 0.7252719 0.5458016 3.012803 2 Spring 1.237781 0.7339281 0.2097657 2.657602 3 Summer 4.854856 2.7684078 0.9625347 10.261749 4 Winter 4.072874 2.8468510 0.5423747 9.202545
- Generate a summary figure
mcmc = quinn.mcmcpack wch = c(which(colnames(mcmc)=='(Intercept)'),grep("^SEASON|^DENSITY", colnames(mcmc))) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
SEASON DENSITY estimate std.error conf.low conf.high 1 Autumn High 4.2252128 0.4241621 3.3943276 5.054871 2 Spring High 3.0155882 0.4300957 2.1510195 3.842651 3 Summer High 6.8669721 0.4250116 5.9918723 7.668388 4 Winter High 2.2714233 0.4223715 1.4241974 3.077524 5 Autumn Low 4.2579776 0.5156992 3.2170528 5.236245 6 Spring Low 3.2931520 0.4671794 2.3578430 4.197222 7 Summer Low 4.6496953 0.4312228 3.8158781 5.497845 8 Winter Low 0.9498468 0.6018597 -0.1876365 2.176258
## Notice that the lower confidence interval for ## Winter Low is less than 0 ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits~(sqrt(x))))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# We could also backtransforms # (issues associated with doing so from root # transformed data notwithstanding) fit=(coefs %*% t(Xmat))^2 newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# Whilst not massively obvious, the lower # confidence interval for Winter Low density is # artificially shrunk due to the effect of # squaring a negative value
mcmc = quinn.r2jags$BUGSoutput$sims.matrix wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
SEASON DENSITY estimate std.error conf.low conf.high 1 Autumn High 4.2275095 0.4300189 3.3871678 5.062626 2 Spring High 3.0240854 0.4324205 2.1670957 3.870135 3 Summer High 6.8736315 0.4329774 6.0495516 7.753169 4 Winter High 2.2735707 0.4304137 1.4140156 3.109802 5 Autumn Low 4.2653552 0.5329208 3.2198356 5.314907 6 Spring Low 3.2953648 0.4794080 2.3235718 4.213940 7 Summer Low 4.6544860 0.4358497 3.7833967 5.492240 8 Winter Low 0.9455688 0.6150566 -0.2309767 2.194197
## Notice that the lower confidence interval for ## Winter Low is less than 0 ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits~(sqrt(x))))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# We could also backtransforms # (issues associated with doing so from root # transformed data notwithstanding) fit=(coefs %*% t(Xmat))^2 newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# Whilst not massively obvious, the lower # confidence interval for Winter Low density is # artificially shrunk due to the effect of # squaring a negative value
mcmc = as.matrix(quinn.rstan) wch = grep("^beta", colnames(mcmc)) ## Calculate the fitted values newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) Xmat = model.matrix(~SEASON*DENSITY,newdata) coefs = mcmc[,wch] fit=coefs %*% t(Xmat) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) newdata
SEASON DENSITY estimate std.error conf.low conf.high 1 Autumn High 4.2489438 0.4254124 3.4227812 5.080433 2 Spring High 3.0077453 0.4323145 2.2043674 3.871054 3 Summer High 6.8562278 0.4254240 5.9969017 7.654856 4 Winter High 2.2680858 0.4350191 1.4018683 3.080181 5 Autumn Low 4.2412038 0.5159389 3.1798805 5.171333 6 Spring Low 3.2923145 0.4644636 2.4231264 4.282321 7 Summer Low 4.6547149 0.4319690 3.8136754 5.433789 8 Winter Low 0.9472953 0.6059751 -0.2413013 2.108698
## Notice that the lower confidence interval for ## Winter Low is less than 0 ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits~(sqrt(x))))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# We could also backtransforms # (issues associated with doing so from root # transformed data notwithstanding) fit=(coefs %*% t(Xmat))^2 newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# Whilst not massively obvious, the lower # confidence interval for Winter Low density is # artificially shrunk due to the effect of # squaring a negative value
newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) fit = posterior_linpred(quinn.rstanarm, newdata = newdata) newdata = newdata %>% cbind(tidyMCMC(as.mcmc(fit),conf.int = TRUE,conf.method = "HPDinterval")) newdata
SEASON DENSITY estimate std.error conf.low conf.high 1 Autumn High 4.294462 0.3781257 3.60088538 5.078098 2 Spring High 3.100023 0.4164097 2.27168435 3.918332 3 Summer High 6.645078 0.4141896 5.79403526 7.437191 4 Winter High 2.335080 0.4014309 1.52534364 3.103879 5 Autumn Low 4.091354 0.4685777 3.11503025 5.001308 6 Spring Low 3.265627 0.4699615 2.33196121 4.197822 7 Summer Low 4.748520 0.4265861 3.89085842 5.550152 8 Winter Low 1.080742 0.5927495 -0.05760336 2.246805
## Notice that the lower confidence interval for ## Winter Low is less than 0 ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits~(sqrt(x))))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# We could also backtransforms # (issues associated with doing so from root # transformed data notwithstanding) mcmc = as.matrix(quinn.rstanarm) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SEASON|^DENSITY',colnames(mcmc))) coefs = mcmc[,wch] fit=(coefs %*% t(Xmat))^2 newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# Whilst not massively obvious, the lower # confidence interval for Winter Low density is # artificially shrunk due to the effect of # squaring a negative value
## The simple way plot(marginal_effects(quinn.brms))



## OR eff=marginal_effects(quinn.brms) ggplot(eff[['SEASON:DENSITY']], aes(y=estimate__, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=lower__, ymax=upper__), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits~(sqrt(x))))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# We could also backtransforms # (issues associated with doing so from root # transformed data notwithstanding) mcmc = as.matrix(quinn.brms) wch = grep('^b_',colnames(mcmc)) coefs = mcmc[,wch] fit=(coefs %*% t(Xmat))^2 newdata = expand.grid(SEASON=levels(quinn$SEASON), DENSITY=levels(quinn$DENSITY)) newdata = newdata %>% cbind(tidyMCMC(fit, conf.int=TRUE, conf.method='HPDinterval')) ggplot(newdata, aes(y=estimate, x=SEASON, fill=DENSITY)) + geom_blank() + geom_line(aes(x=as.numeric(SEASON), linetype=DENSITY), position=position_dodge(width=0.2)) + geom_linerange(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(width=0.2))+ geom_point(aes(shape=DENSITY), size=3, position=position_dodge(width=0.2))+ scale_y_continuous(expression(Number~of~recruits))+ scale_x_discrete('SEASON')+ scale_shape_manual('DENSITY',values=c(21,16))+ scale_fill_manual('DENSITY',values=c('white','black'))+ scale_linetype_manual('DENSITY',values=c('solid','dashed'))+ theme_classic() + theme(legend.justification=c(1,1), legend.position=c(1,1), axis.title.y=element_text(vjust=2, size=rel(1.25)), axis.title.x=element_text(vjust=-2, size=rel(1.25)), plot.margin=unit(c(0.5,0.5,2,2), 'lines'), legend.key.width=unit(1,'cm'))

# Whilst not massively obvious, the lower # confidence interval for Winter Low density is # artificially shrunk due to the effect of # squaring a negative value
- Explore finite-population standard deviations
mcmc = quinn.mcmcpack Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = attr(Xmat, 'assign') wch
[1] 0 1 1 1 2 3 3 3
# Get the rowwise standard deviations between effects parameters sd.SEASON = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.DENSITY = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SEASON|^DENSITY',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,quinn$SQRTRECRUITS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SEASON, sd.DENSITY, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 2.0458591 0.24399853 1.5713229883 2.5271583 2 sd.DENSITY 0.3721277 0.28736102 0.0000249148 0.9274377 3 sd.Int 1.0546492 0.30297126 0.4743949616 1.6452895 4 sd.resid 1.0142058 0.05421245 0.9293554586 1.1183462
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 46.507127 4.950428 3.635905e+01 55.13205 2 sd.DENSITY 7.045301 5.452521 6.341264e-04 18.14570 3 sd.Int 23.768224 4.813691 1.348344e+01 31.75094 4 sd.resid 22.680436 2.711154 1.780369e+01 28.34489
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = quinn.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SEASON = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.DENSITY = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,quinn$SQRTRECRUITS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SEASON, sd.DENSITY, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 2.0476087 0.24732642 1.5663547655 2.5384537 2 sd.DENSITY 0.3863201 0.29508764 0.0002549547 0.9519804 3 sd.Int 1.0612923 0.31022276 0.4733156490 1.6792146 4 sd.resid 1.0178450 0.05586504 0.9310637914 1.1248443
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 46.279973 5.043371 35.208918587 54.36125 2 sd.DENSITY 7.340608 5.545821 0.005864635 18.38700 3 sd.Int 23.780605 4.881884 13.179298142 31.65971 4 sd.resid 22.641378 2.737040 17.696588483 28.39004
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(quinn.rstan) Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SEASON = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.DENSITY = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^beta',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,quinn$SQRTRECRUITS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SEASON, sd.DENSITY, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 2.0436502 0.24992687 1.5987749360 2.5320684 2 sd.DENSITY 0.3692038 0.28777371 0.0003072288 0.9240513 3 sd.Int 1.0391082 0.29672574 0.4980013885 1.6747557 4 sd.resid 1.0148922 0.05512689 0.9350503790 1.1250084
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 46.681985 4.974681 36.132920311 54.82294 2 sd.DENSITY 7.009301 5.586518 0.008148223 18.54548 3 sd.Int 23.566690 4.863698 13.338071480 31.58339 4 sd.resid 22.698354 2.733369 18.161911084 28.36202
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(quinn.rstanarm) Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SEASON = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.DENSITY = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SEASON|^DENSITY',colnames(mcmc))) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,quinn$SQRTRECRUITS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SEASON, sd.DENSITY, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 1.9098388 0.23629616 1.456012e+00 2.3773175 2 sd.DENSITY 0.3248635 0.24596846 7.510478e-05 0.8047007 3 sd.Int 0.8486887 0.24904106 3.598045e-01 1.3469656 4 sd.resid 1.0133378 0.05291751 9.321180e-01 1.1168406
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 47.320279 4.535517 37.69044017 55.02143 2 sd.DENSITY 6.706523 5.392505 0.00920687 17.96551 3 sd.Int 20.920268 4.909256 10.71234898 29.80968 4 sd.resid 24.712207 2.956246 19.46343811 30.74403
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

mcmc = as.matrix(quinn.brms) Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = attr(Xmat, 'assign') # Get the rowwise standard deviations between effects parameters sd.SEASON = apply(cbind(0,mcmc[,which(wch %in% c(1))]),1,sd) sd.DENSITY = apply(cbind(0,mcmc[,which(wch %in% c(2))]),1,sd) sd.Int = apply(cbind(0,0,0,0,mcmc[,which(wch %in% c(3))]),1,sd) # generate a model matrix newdata = data ## get median parameter estimates wch = grep('^b_',colnames(mcmc)) coefs = mcmc[, wch] fit = coefs %*% t(Xmat) resid = sweep(fit,2,quinn$SQRTRECRUITS,'-') sd.resid = apply(resid,1,sd) sd.all = cbind(sd.SEASON, sd.DENSITY, sd.Int, sd.resid) (fpsd = tidyMCMC(sd.all, conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 1.6778093 0.22358321 1.253933e+00 2.1260798 2 sd.DENSITY 0.3224921 0.23195720 6.486431e-07 0.7694802 3 sd.Int 0.6113971 0.20386152 1.992382e-01 0.9937867 4 sd.resid 1.0358146 0.06059855 9.334732e-01 1.1517172
#OR expressed as a percentage (fpsd.p = tidyMCMC(100*sd.all/rowSums(sd.all), estimate.method='median', conf.int=TRUE, conf.method='HPDinterval'))
term estimate std.error conf.low conf.high 1 sd.SEASON 46.452154 4.597217 3.690050e+01 54.15032 2 sd.DENSITY 7.768399 5.751150 1.871847e-05 19.27625 3 sd.Int 16.801591 4.963831 6.694434e+00 26.18510 4 sd.resid 28.246663 3.753022 2.215555e+01 36.19455
fpsd = fpsd %>% mutate(term = factor(term, levels=unique(.$term))) fpsd.p = fpsd.p %>% mutate(term = factor(term, levels=unique(.$term))) ## we can even plot this as a Bayesian ANOVA table ggplot(fpsd, aes(y=estimate, x=term)) + geom_hline(yintercept=0, linetype='dashed') + geom_pointrange(aes(ymin=conf.low, ymax=conf.high)) + geom_text(aes(label=sprintf('%.2f%%', fpsd.p$estimate), vjust=-1)) + scale_y_continuous('Finite population standard deviation') + scale_x_discrete()+ coord_flip() + theme_classic()

- Estimate a psuedo-$R^2$
library(broom) mcmc <- quinn.mcmcpack Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = c(which(colnames(mcmc)=='(Intercept)'), grep('^SEASON|^DENSITY', colnames(mcmc))) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, quinn$SQRTRECRUITS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.7240554 0.04256129 0.6396949 0.7890885
#for comparison with frequentist summary(lm(SQRTRECRUITS ~ SEASON*DENSITY, quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08library(broom) mcmc <- quinn.r2jags$BUGSoutput$sims.matrix Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = grep('^beta', colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, quinn$SQRTRECRUITS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.7233417 0.04363577 0.637284 0.790102
#for comparison with frequentist summary(lm(SQRTRECRUITS ~ SEASON*DENSITY, quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08library(broom) mcmc <- as.matrix(quinn.rstan) Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = grep('^beta', colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, quinn$SQRTRECRUITS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.7236663 0.04337963 0.6414205 0.7903614
#for comparison with frequentist summary(lm(SQRTRECRUITS ~ SEASON*DENSITY, quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08library(broom) mcmc <- as.matrix(quinn.rstanarm) Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = c(which(colnames(mcmc) == '(Intercept)'), grep('^SEASON|^DENSITY',colnames(mcmc))) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, quinn$SQRTRECRUITS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.7014785 0.05259404 0.5959346 0.7831196
#for comparison with frequentist summary(lm(SQRTRECRUITS ~ SEASON*DENSITY, quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08library(broom) mcmc <- as.matrix(quinn.brms) Xmat = model.matrix(~SEASON*DENSITY, quinn) wch = grep('^b_',colnames(mcmc)) coefs = mcmc[,wch] fit = coefs %*% t(Xmat) resid = sweep(fit, 2, quinn$SQRTRECRUITS, "-") var_f = apply(fit,1,var) var_e = apply(resid,1, var) R2 = var_f/(var_f + var_e) tidyMCMC(as.mcmc(R2), conf.int = TRUE, conf.method = "HPDinterval")
term estimate std.error conf.low conf.high 1 var1 0.6508712 0.06867597 0.5235427 0.7711021
#for comparison with frequentist summary(lm(SQRTRECRUITS ~ SEASON*DENSITY, quinn))
Call: lm(formula = SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) Residuals: Min 1Q Median 3Q Max -2.2298 -0.5977 0.1384 0.5489 1.8856 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.22979 0.41235 10.258 6.06e-12 *** SEASONSpring -1.20984 0.58315 -2.075 0.04566 * SEASONSummer 2.63583 0.58315 4.520 7.14e-05 *** SEASONWinter -1.95739 0.58315 -3.357 0.00195 ** DENSITYLow 0.02996 0.65198 0.046 0.96362 SEASONSpring:DENSITYLow 0.24657 0.89396 0.276 0.78436 SEASONSummer:DENSITYLow -2.24336 0.87473 -2.565 0.01492 * SEASONWinter:DENSITYLow -1.35956 0.96705 -1.406 0.16883 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 1.01 on 34 degrees of freedom Multiple R-squared: 0.7533, Adjusted R-squared: 0.7025 F-statistic: 14.83 on 7 and 34 DF, p-value: 1.097e-08
