Workshop 6 - Factorial Analysis of Variance
5 July 2011
Factorial ANOVA references
- Logan (2010) - Chpt 12
- Quinn & Keough (2002) - Chpt 9
Very basic overview of Two-Factor ANOVA
Nested ANOVA - one between factor
In an unusually detailed preparation for an Environmental Effects Statement for a proposed discharge of dairy wastes into the Curdies River, in western Victoria, a team of stream ecologists wanted to describe the basic patterns of variation in a stream invertebrate thought to be sensitive to nutrient enrichment. As an indicator species, they focused on a small flatworm, Dugesia, and started by sampling populations of this worm at a range of scales. They sampled in two seasons, representing different flow regimes of the river - Winter and Summer. Within each season, they sampled three randomly chosen (well, haphazardly, because sites are nearly always chosen to be close to road access) sites. A total of six sites in all were visited, 3 in each season. At each site, they sampled six stones, and counted the number of flatworms on each stone.
Download Curdies data set
| Format of curdies.csv data files | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||
> curdies <- read.table("../downloads/data/curdies.csv", header = T, + sep = ",", strip.white = T) > head(curdies)SEASON SITE DUGESIA S4DUGES 1 WINTER 1 0.6477 0.8971 2 WINTER 1 6.0962 1.5713 3 WINTER 1 1.3106 1.0700 4 WINTER 1 1.7253 1.1461 5 WINTER 1 1.4594 1.0991 6 WINTER 1 1.0576 1.0141
> curdies$SITE <- as.factor(curdies$SITE)
Notice the data set - each of the nested factors is labelled differently - there can be no replicate for the random (nesting) factor.
- H0 Effect 1:
- H0 Effect 2:
| Assumption | Diagnostic/Risk Minimization |
|---|---|
| I. | |
| II. | |
| III. |
> library(nlme) > curdies.ag <- gsummary(curdies, form = ~SEASON/SITE, mean) > curdies.agSEASON SITE DUGESIA S4DUGES WINTER/1 WINTER 1 2.0494 1.1329 WINTER/2 WINTER 2 4.1819 1.2719 WINTER/3 WINTER 3 0.6782 0.8679 SUMMER/4 SUMMER 4 0.4191 0.3508 SUMMER/5 SUMMER 5 0.2291 0.1805 SUMMER/6 SUMMER 6 0.1942 0.3811
> boxplot(DUGESIA ~ SEASON, curdies.ag)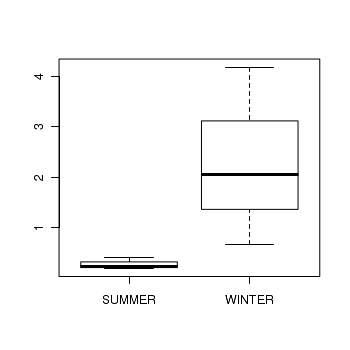
If so, assess whether a transformation will address the violations (HINT) and then make the appropriate corrections
> curdies$S4DUGES <- sqrt(sqrt(curdies$DUGESIA)) > curdies.ag <- gsummary(curdies, form = ~SEASON/SITE, mean) > boxplot(S4DUGES ~ SEASON, curdies.ag)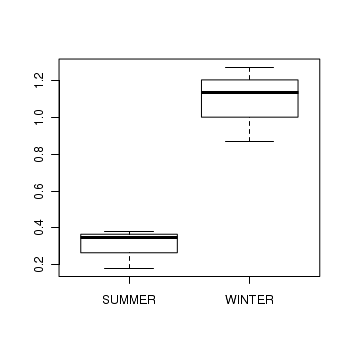
| Effect | Nominator (Mean Sq, df) | Denominator (Mean Sq, df) |
|---|---|---|
| SEASON | ||
| SITE |
S4DUGES = SEASON + SITE + CONSTANT
using a nested ANOVA (HINT). Fill (HINT) out the table below, make sure that you have treated SITE as a random factor when compiling the overall results.
> curdies.aov <- aov(S4DUGES ~ SEASON + Error(SITE), data = curdies) > summary(curdies.aov)Error: SITE Df Sum Sq Mean Sq F value Pr(>F) SEASON 1 5.57 5.57 34.5 0.0042 ** Residuals 4 0.65 0.16 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Error: Within Df Sum Sq Mean Sq F value Pr(>F) Residuals 30 4.56 0.152> summary(lm(curdies.aov))Call: lm(formula = curdies.aov) Residuals: Min 1Q Median 3Q Max -0.381 -0.262 -0.138 0.165 0.902 Coefficients: (1 not defined because of singularities) Estimate Std. Error t value Pr(>|t|) (Intercept) 0.3811 0.1591 2.40 0.0230 * SEASONWINTER 0.7518 0.2250 3.34 0.0022 ** SITE2 0.1389 0.2250 0.62 0.5416 SITE3 -0.2651 0.2250 -1.18 0.2480 SITE4 -0.0303 0.2250 -0.13 0.8938 SITE5 -0.2007 0.2250 -0.89 0.3795 SITE6 NA NA NA NA --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.39 on 30 degrees of freedom Multiple R-squared: 0.577, Adjusted R-squared: 0.507 F-statistic: 8.19 on 5 and 30 DF, p-value: 5.72e-05> confint(lm(curdies.aov))2.5 % 97.5 % (Intercept) 0.05622 0.7060 SEASONWINTER 0.29235 1.2113 SITE2 -0.32055 0.5984 SITE3 -0.72455 0.1944 SITE4 -0.48978 0.4292 SITE5 -0.66013 0.2588 SITE6 NA NA
| Source of variation | df | Mean Sq | F-ratio | P-value |
|---|---|---|---|---|
| SEASON | ||||
| SITE | ||||
| Residuals |
| Estimate | Mean | Lower 95% CI | Upper 95% CI |
|---|---|---|---|
| Summer | |||
| Effect size (Winter-Summer) |
Normally, we are not interested in formally testing the effect of the nested factor to get the correct F test for the nested factor (SITE), examine a representation of the anova table of the fitted linear model that assumes all factors are fixed (HINT)
> library(nlme) > VarCorr(lme(S4DUGES ~ 1, random = ~1 | SEASON/SITE, curdies))Variance StdDev SEASON = pdLogChol(1) (Intercept) 0.300523 0.54820 SITE = pdLogChol(1) (Intercept) 0.001607 0.04009 Residual 0.151851 0.38968
- What influences the abundance of Dugesia
- Where best to focus sampling effort to maximize statistical power?
> opar <- par(mar = c(5, 5, 1, 1)) > means <- tapply(curdies.ag$DUGESIA, curdies.ag$SEASON, mean) > sds <- tapply(curdies.ag$DUGESIA, curdies.ag$SEASON, sd) > lens <- tapply(curdies.ag$DUGESIA, curdies.ag$SEASON, length) > ses <- sds/sqrt(lens) > xs <- barplot(means, beside = T, ann = F, axes = F, ylim = c(0, max(means + + ses)), axisnames = F) > arrows(xs, means - ses, xs, means + ses, code = 3, length = 0.05, + ang = 90) > axis(1, at = xs, lab = c("Summer", "Winter")) > mtext("Season", 1, line = 3, cex = 1.25) > axis(2, las = 1) > mtext("Mean number of Dugesia per stone", 2, line = 3, cex = 1.25) > box(bty = "l")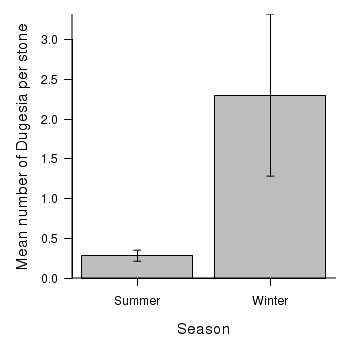
Exercise 2 - Two factor ANOVA
A biologist studying starlings wanted to know whether the mean mass of starlings differed according to different roosting situations. She was also interested in whether the mean mass of starlings altered over winter (Northern hemisphere) and whether the patterns amongst roosting situations were consistent throughout winter, therefore starlings were captured at the start (November) and end of winter (January). Ten starlings were captured from each roosting situation in each season, so in total, 80 birds were captured and weighed.
Download Starling data set
| Format of starling.csv data files | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Open the starling data file.
> starling <- read.table("../downloads/data/starling.csv", header = T, + sep = ",", strip.white = T) > head(starling)SITUATION MONTH MASS GROUP 1 S1 November 78 S1Nov 2 S1 November 88 S1Nov 3 S1 November 87 S1Nov 4 S1 November 88 S1Nov 5 S1 November 83 S1Nov 6 S1 November 82 S1Nov
> boxplot(MASS ~ SITUATION * MONTH, data = starling)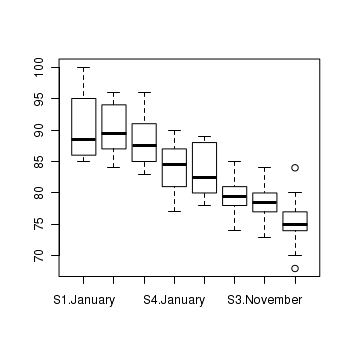 > means <- with(starling, tapply(MASS, list(SITUATION, MONTH), mean)) > vars <- with(starling, tapply(MASS, list(SITUATION, MONTH), var)) > plot(means, vars, pch = 16)
> means <- with(starling, tapply(MASS, list(SITUATION, MONTH), mean)) > vars <- with(starling, tapply(MASS, list(SITUATION, MONTH), var)) > plot(means, vars, pch = 16)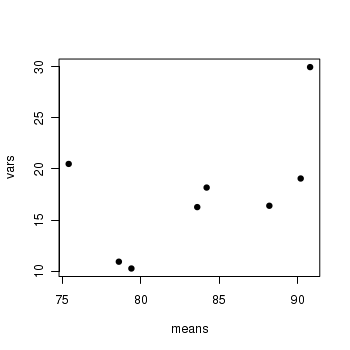
- Is there any evidence that one or more of the assumptions are likely to be violated? (Y or N)
- Is the proposed model balanced?
(Y or N)
> replications(MASS ~ SITUATION * MONTH, data = starling)SITUATION MONTH SITUATION:MONTH 20 40 10> !is.list(replications(MASS ~ SITUATION * MONTH, data = starling))[1] TRUE
> starling.lm <- lm(MASS ~ SITUATION * MONTH, data = starling) > # setup a 2x6 plotting device with small margins > par(mfrow = c(2, 1), oma = c(0, 0, 2, 0)) > plot(starling.lm, ask = F, which = 1:2)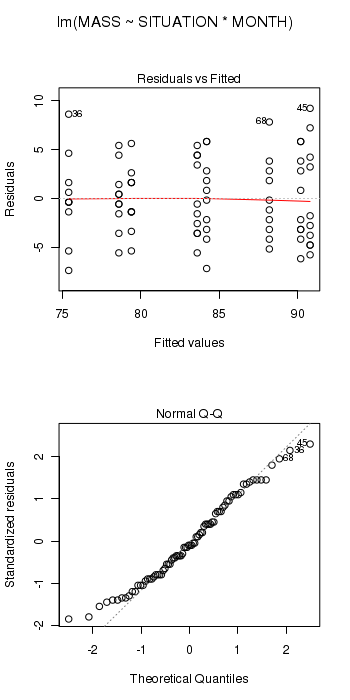
Examine the ANOVA table
> summary(starling.lm)Call: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Residuals: Min 1Q Median 3Q Max -7.4 -3.2 -0.4 2.9 9.2 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 90.80 1.33 68.26 < 2e-16 *** SITUATIONS2 -0.60 1.88 -0.32 0.75069 SITUATIONS3 -2.60 1.88 -1.38 0.17121 SITUATIONS4 -6.60 1.88 -3.51 0.00078 *** MONTHNovember -7.20 1.88 -3.83 0.00027 *** SITUATIONS2:MONTHNovember -3.60 2.66 -1.35 0.18023 SITUATIONS3:MONTHNovember -2.40 2.66 -0.90 0.37000 SITUATIONS4:MONTHNovember -1.60 2.66 -0.60 0.54946 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 4.21 on 72 degrees of freedom Multiple R-squared: 0.64, Adjusted R-squared: 0.605 F-statistic: 18.3 on 7 and 72 DF, p-value: 9.55e-14> anova(starling.lm)Analysis of Variance Table Response: MASS Df Sum Sq Mean Sq F value Pr(>F) SITUATION 3 574 191 10.82 6.0e-06 *** MONTH 1 1656 1656 93.60 1.2e-14 *** SITUATION:MONTH 3 34 11 0.64 0.59 Residuals 72 1274 18 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
| Source of Variation | SS | df | MS | F-ratio | Pvalue |
|---|---|---|---|---|---|
| SITUATION | |||||
| MONTH | |||||
| SITUATION : MONTH | |||||
| Residual (within groups) |
> library(car) > with(starling, interaction.plot(SITUATION, MONTH, MASS))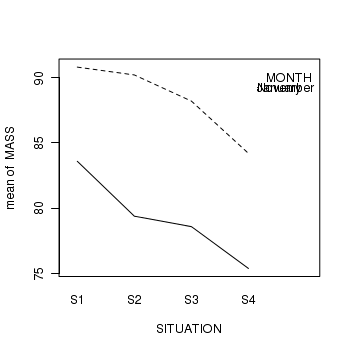
In the classical frequentist regime, many at this point would advocate dropping the interaction term from the model (p-value for the interaction is greater than 0.25). This term is not soaking up much of the residual and yet is soaking up 3 degrees of freedom. The figure also indicates that situation and month are likely to operate additively.
> library(multcomp) > summary(glht(starling.lm, linfct = mcp(SITUATION = "Tukey")))Simultaneous Tests for General Linear Hypotheses Multiple Comparisons of Means: Tukey Contrasts Fit: lm(formula = MASS ~ SITUATION * MONTH, data = starling) Linear Hypotheses: Estimate Std. Error t value Pr(>|t|) S2 - S1 == 0 -0.60 1.88 -0.32 0.9887 S3 - S1 == 0 -2.60 1.88 -1.38 0.5146 S4 - S1 == 0 -6.60 1.88 -3.51 0.0044 ** S3 - S2 == 0 -2.00 1.88 -1.06 0.7129 S4 - S2 == 0 -6.00 1.88 -3.19 0.0111 * S4 - S3 == 0 -4.00 1.88 -2.13 0.1546 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 (Adjusted p values reported -- single-step method)> library(multcomp) > summary(glht(lm(MASS ~ SITUATION + MONTH, data = starling), linfct = mcp(SITUATION = "Tukey")))Simultaneous Tests for General Linear Hypotheses Multiple Comparisons of Means: Tukey Contrasts Fit: lm(formula = MASS ~ SITUATION + MONTH, data = starling) Linear Hypotheses: Estimate Std. Error t value Pr(>|t|) S2 - S1 == 0 -2.40 1.32 -1.82 0.2734 S3 - S1 == 0 -3.80 1.32 -2.88 0.0264 * S4 - S1 == 0 -7.40 1.32 -5.60 <0.001 *** S3 - S2 == 0 -1.40 1.32 -1.06 0.7147 S4 - S2 == 0 -5.00 1.32 -3.79 0.0017 ** S4 - S3 == 0 -3.60 1.32 -2.73 0.0392 * --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 (Adjusted p values reported -- single-step method)> confint(glht(lm(MASS ~ SITUATION + MONTH, data = starling), linfct = mcp(SITUATION = "Tukey")))Simultaneous Confidence Intervals Multiple Comparisons of Means: Tukey Contrasts Fit: lm(formula = MASS ~ SITUATION + MONTH, data = starling) Quantile = 2.628 95% family-wise confidence level Linear Hypotheses: Estimate lwr upr S2 - S1 == 0 -2.400 -5.870 1.070 S3 - S1 == 0 -3.800 -7.270 -0.330 S4 - S1 == 0 -7.400 -10.870 -3.930 S3 - S2 == 0 -1.400 -4.870 2.070 S4 - S2 == 0 -5.000 -8.470 -1.530 S4 - S3 == 0 -3.600 -7.070 -0.130
| Comparison | Multiplicative | Additive | ||||||
|---|---|---|---|---|---|---|---|---|
| Est | P | Lwr | Upr | Est | P | Lwr | Upr | |
| Situation 2 vs Situation 1 | ||||||||
| Situation 3 vs Situation 1 | ||||||||
| Situation 4 vs Situation 1 | ||||||||
| Situation 3 vs Situation 2 | ||||||||
| Situation 4 vs Situation 2 | ||||||||
| Situation 4 vs Situation 3 | ||||||||
> starling.lm2 <- lm(MASS ~ SITUATION + MONTH, data = starling) > cbind(coef(starling.lm2), confint(starling.lm2))2.5 % 97.5 % (Intercept) 91.75 89.670 93.830 SITUATIONS2 -2.40 -5.031 0.231 SITUATIONS3 -3.80 -6.431 -1.169 SITUATIONS4 -7.40 -10.031 -4.769 MONTHNovember -9.10 -10.960 -7.240> contrasts(starling$SITUATION) <- contr.sum
| Estimate | Mean | Lower 95% CI | Upper 95% CI |
|---|---|---|---|
| December Situation 1 | |||
| Effect size (Dec:Sit2 - Dec:Sit1) | |||
| Effect size (Dec:Sit3 - Dec:Sit1) | |||
| Effect size (Dec:Sit4 - Dec:Sit1) | |||
| Effect size (Nov:Sit1 - Dec:Sit1) |
> opar <- par(mar = c(5, 5, 1, 1)) > star.means <- with(starling, tapply(MASS, list(MONTH, SITUATION), + mean)) > star.sds <- with(starling, tapply(MASS, list(MONTH, SITUATION), sd)) > star.len <- with(starling, tapply(MASS, list(MONTH, SITUATION), length)) > star.ses <- star.sds/sqrt(star.len) > xs <- barplot(star.means, ylim = range(starling$MASS), beside = T, + axes = F, ann = F, axisnames = F, xpd = F, axis.lty = 2, col = c(0, 1)) > arrows(xs, star.means, xs, star.means + star.ses, code = 2, length = 0.05, + ang = 90) > axis(2, las = 1) > axis(1, at = apply(xs, 2, median), lab = c("Situation 1", "Situation 2", + "Situation 3", "Situation 4")) > mtext(2, text = "Mass (g) of starlings", line = 3, cex = 1.25) > legend("topright", leg = c("January", "November"), fill = c(0, 1), + col = c(0, 1), bty = "n", cex = 1) > box(bty = "l")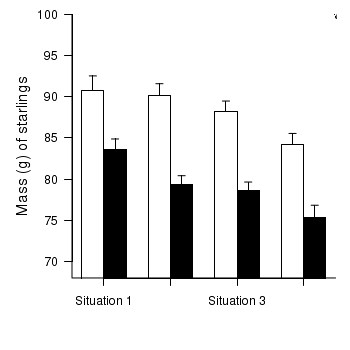 > par(opar)
> par(opar)
Exercise 3 - Two factor ANOVA - Type III SS
Here is a modified example from Quinn and Keough (2002). Stehman and Meredith (1995) present data from an experiment that was set up to test the hypothesis that healthy spruce seedlings break bud sooner than diseased spruce seedlings. There were 2 factors: pH (3 levels: 3, 5.5, 7) and HEALTH (2 levels: healthy, diseased). The dependent variable was the average (from 5 buds) bud emergence rating (BRATING) on each seedling. The sample size varied for each combination of pH and health, ranging from 7 to 23 seedlings. With two factors, this experiment should be analyzed with a 2 factor (2 x 3) ANOVA.
Download Stehman data set
| Format of stehman.csv data files | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Open the stehman data file.
> stehman <- read.table("../downloads/data/stehman.csv", header = T, + sep = ",", strip.white = T) > head(stehman)PH HEALTH GROUP BRATING 1 3 D D3 0.0 2 3 D D3 0.8 3 3 D D3 0.8 4 3 D D3 0.8 5 3 D D3 0.8 6 3 D D3 0.8
> stehman$PH <- as.factor(stehman$PH)
> boxplot(BRATING ~ HEALTH * PH, data = stehman)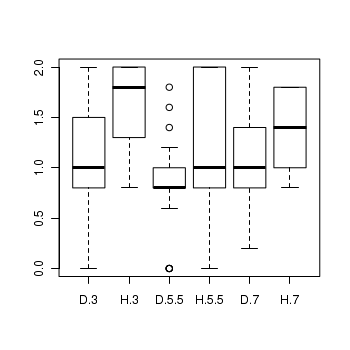 > means <- with(stehman, tapply(BRATING, list(HEALTH, PH), mean)) > vars <- with(stehman, tapply(BRATING, list(HEALTH, PH), var)) > plot(means, vars, pch = 16)
> means <- with(stehman, tapply(BRATING, list(HEALTH, PH), mean)) > vars <- with(stehman, tapply(BRATING, list(HEALTH, PH), var)) > plot(means, vars, pch = 16)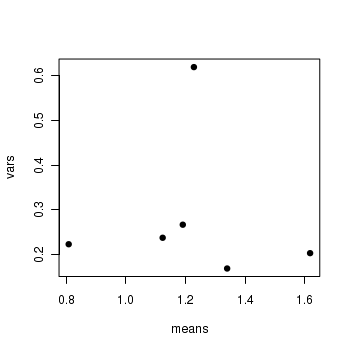
- Is there any evidence that one or more of the assumptions are likely to be violated? (Y or N)
- Is the proposed model balanced?
(Y or N)
> replications(BRATING ~ HEALTH * PH, data = stehman)$HEALTH HEALTH D H 67 28 $PH PH 3 5.5 7 34 30 31 $`HEALTH:PH` PH HEALTH 3 5.5 7 D 23 23 21 H 11 7 10> !is.list(replications(BRATING ~ HEALTH * PH, data = stehman))[1] FALSE
As the model is not balanced, we will likely want to examine the ANOVA table based on Type III (marginal) Sums of Squares. In preparation for doing so, we must define something other than treatment contrasts for the factors.
> contrasts(stehman$HEALTH) <- contr.sum > contrasts(stehman$PH) <- contr.sum
> stehman.lm <- lm(BRATING ~ HEALTH * PH, data = stehman) > library(car) > Anova(stehman.lm, type = "III")Anova Table (Type III tests) Response: BRATING Sum Sq Df F value Pr(>F) (Intercept) 114.1 1 444.87 <2e-16 *** HEALTH 2.4 1 9.40 0.0029 ** PH 1.9 2 3.63 0.0306 * HEALTH:PH 0.2 2 0.37 0.6897 Residuals 22.8 89 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
| Source of Variation | SS | df | MS | F-ratio | Pvalue |
|---|---|---|---|---|---|
| PH | |||||
| HEALTH | |||||
| PH : HEALTH | |||||
| Residual (within groups) |
> library(car) > with(stehman, interaction.plot(PH, HEALTH, BRATING))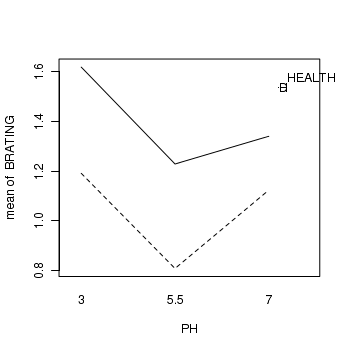
> library(multcomp) > summary(glht(stehman.lm, linfct = mcp(PH = "Tukey")))Simultaneous Tests for General Linear Hypotheses Multiple Comparisons of Means: Tukey Contrasts Fit: lm(formula = BRATING ~ HEALTH * PH, data = stehman) Linear Hypotheses: Estimate Std. Error t value Pr(>|t|) 5.5 - 3 == 0 -0.386 0.143 -2.69 0.023 * 7 - 3 == 0 -0.173 0.134 -1.29 0.407 7 - 5.5 == 0 0.213 0.146 1.46 0.316 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 (Adjusted p values reported -- single-step method)
pH 3 vs pH 5.5
pH 3 vs pH 7
pH 5.5 vs pH 7
The above analysis reflects a very classic approach to investigating the effects of PH and HEALTH via NHST (null hypothesis testing). Many argue that a more modern/valid approach is to:
- Abandon hypothesis testing
- Estimate effect sizes and associated effect sizes
- As PH is an ordered factor, this is arguably better modelled via polynomial contrasts
> stehman.lm2 <- lm(BRATING ~ PH * HEALTH, data = stehman, contrasts = list(PH = contr.poly(3, + scores = c(3, 5.5, 7)), HEALTH = contr.treatment)) > summary(stehman.lm2)Call: lm(formula = BRATING ~ PH * HEALTH, data = stehman, contrasts = list(PH = contr.poly(3, scores = c(3, 5.5, 7)), HEALTH = contr.treatment)) Residuals: Min 1Q Median 3Q Max -1.2286 -0.3238 -0.0087 0.3818 0.9913 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 1.0413 0.0619 16.81 <2e-16 *** PH.L -0.0879 0.1077 -0.82 0.4162 PH.Q 0.2751 0.1069 2.57 0.0117 * HEALTH2 0.3543 0.1155 3.07 0.0029 ** PH.L:HEALTH2 -0.1360 0.1899 -0.72 0.4758 PH.Q:HEALTH2 -0.1008 0.2099 -0.48 0.6323 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 0.506 on 89 degrees of freedom Multiple R-squared: 0.196, Adjusted R-squared: 0.15 F-statistic: 4.33 on 5 and 89 DF, p-value: 0.00143> confint(stehman.lm2)2.5 % 97.5 % (Intercept) 0.91821 1.1643 PH.L -0.30184 0.1260 PH.Q 0.06273 0.4875 HEALTH2 0.12475 0.5839 PH.L:HEALTH2 -0.51324 0.2413 PH.Q:HEALTH2 -0.51773 0.3162
- Healthy spruce trees have a higher bud emergence rating than diseased (ES=0.35 CI=0.12-0.58)
- The bud emergence rating follows a quadratic change with PH
Exercise 4 - Two factor ANOVA
An ecologist studying a rocky shore at Phillip Island, in southeastern Australia, was interested in how clumps of intertidal mussels are maintained. In particular, he wanted to know how densities of adult mussels affected recruitment of young individuals from the plankton. As with most marine invertebrates, recruitment is highly patchy in time, so he expected to find seasonal variation, and the interaction between season and density - whether effects of adult mussel density vary across seasons - was the aspect of most interest.
The data were collected from four seasons, and with two densities of adult mussels. The experiment consisted of clumps of adult mussels attached to the rocks. These clumps were then brought back to the laboratory, and the number of baby mussels recorded. There were 3-6 replicate clumps for each density and season combination.
Download Quinn data set
| Format of quinn.csv data files | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
Open the quinn data file.
> quinn <- read.table("../downloads/data/quinn.csv", header = T, sep = ",", + strip.white = T) > head(quinn)SEASON DENSITY RECRUITS SQRTRECRUITS GROUP 1 Spring Low 15 3.873 SpringLow 2 Spring Low 10 3.162 SpringLow 3 Spring Low 13 3.606 SpringLow 4 Spring Low 13 3.606 SpringLow 5 Spring Low 5 2.236 SpringLow 6 Spring High 11 3.317 SpringHigh
Confirm the need for a square root transformation, by examining boxplots
and mean vs variance plots for both raw and transformed data. Note that square root transformation was selected because the data were counts (count data often includes values of zero - cannot compute log of zero).> par(mfrow = c(2, 2)) > boxplot(RECRUITS ~ SEASON * DENSITY, data = quinn) > means <- with(quinn, tapply(RECRUITS, list(SEASON, DENSITY), mean)) > vars <- with(quinn, tapply(RECRUITS, list(SEASON, DENSITY), var)) > plot(means, vars, pch = 16) > boxplot(SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) > means <- with(quinn, tapply(SQRTRECRUITS, list(SEASON, DENSITY), + mean)) > vars <- with(quinn, tapply(SQRTRECRUITS, list(SEASON, DENSITY), var)) > plot(means, vars, pch = 16)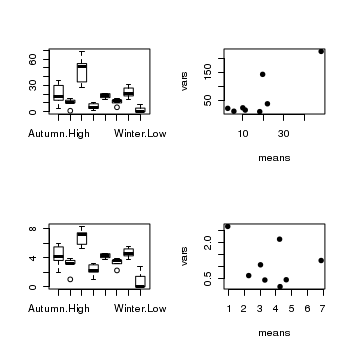
> !is.list(replications(sqrt(RECRUITS) ~ SEASON * DENSITY, data = quinn))[1] FALSE> replications(sqrt(RECRUITS) ~ SEASON * DENSITY, data = quinn)$SEASON SEASON Autumn Spring Summer Winter 10 11 12 9 $DENSITY DENSITY High Low 24 18 $`SEASON:DENSITY` DENSITY SEASON High Low Autumn 6 4 Spring 6 5 Summer 6 6 Winter 6 3> contrasts(quinn$SEASON) <- contr.sum > contrasts(quinn$DENSITY) <- contr.sum
> quinn.lm <- lm(SQRTRECRUITS ~ SEASON * DENSITY, data = quinn) > library(car) > Anova(quinn.lm, type = "III")Anova Table (Type III tests) Response: SQRTRECRUITS Sum Sq Df F value Pr(>F) (Intercept) 540 1 529.04 < 2e-16 *** SEASON 91 3 29.61 1.3e-09 *** DENSITY 6 1 6.35 0.017 * SEASON:DENSITY 11 3 3.71 0.021 * Residuals 35 34 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
| Source of Variation | SS | df | MS | F-ratio | Pvalue |
|---|---|---|---|---|---|
| SEASON | |||||
| DENSITY | |||||
| SEASON : DENSITY | |||||
| Residual (within groups) |
> library(car) > with(quinn, interaction.plot(SEASON, DENSITY, SQRTRECRUITS))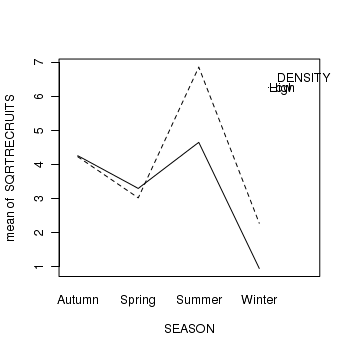
For the current data set, the effect of density is of greatest interest, and thus the former option is the most interesting. Perform the simple main effects anovas.
Download biology package
> library(biology) > AnovaM(quinn.lm.summer <- mainEffects(quinn.lm, at = SEASON == "Summer"), + type = "III")Df Sum Sq Mean Sq F value Pr(>F) INT 6 91.2 15.20 14.9 2.8e-08 *** DENSITY 1 14.7 14.70 14.4 0.00058 *** Residuals 34 34.7 1.02 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1> library(biology) > AnovaM(quinn.lm. <- mainEffects(quinn.lm, at = SEASON == "Autumn"), + type = "III")Df Sum Sq Mean Sq F value Pr(>F) INT 6 105.9 17.65 17.3 4.7e-09 *** DENSITY 1 0.0 0.00 0.0 0.96 Residuals 34 34.7 1.02 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1> AnovaM(quinn.lm. <- mainEffects(quinn.lm, at = SEASON == "Winter"), + type = "III")Df Sum Sq Mean Sq F value Pr(>F) INT 6 102.4 17.06 16.72 7.1e-09 *** DENSITY 1 3.5 3.54 3.47 0.071 . Residuals 34 34.7 1.02 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1> AnovaM(quinn.lm. <- mainEffects(quinn.lm, at = SEASON == "Spring"), + type = "III")Df Sum Sq Mean Sq F value Pr(>F) INT 6 105.7 17.61 17.3 4.8e-09 *** DENSITY 1 0.2 0.21 0.2 0.65 Residuals 34 34.7 1.02 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
- Was the effect of DENSITY on recruitment consistent across all levels of SEASON? (Y or N)
- How would you interpret these results?
