25 Feb 2012
This Tutorial has been thrown together a little hastily and is therefore not very well
organized - sorry!
Graphical features are demonstrated either via tables of properties or as clickable graphics
that reveal the required R code.
Click on a graphic to reveal/toggle the source code or to
navigate to an expanded section.
This tutorial is intended to be viewed sequentially. It
begins with the basic ggplot framework and then
progressively builds up more and more features as default
elements are gradually replaced to yeild more customized graphics.
Having said that, I am going to start with a sort of
showcase of graphics
which should act
as quick navigation to entire sections devoted to the broad
series of graphs related to each of the featured graphics. I have
intentionally titled each graph according to the main
feature it encapsulates rather than any specific functions
that are
used to produce the features as often a single graphic
requires a combination of features and thus functions.
Furthermore, the grammar of graphics specifications are
sufficiently unfamiliar to many that the relationships between the
types of graphical features a researcher wishes to produce and the
specific syntax required to achieve the desired result can
be difficult to recognize.
Each graphic is intended to
encapsulate a broad series of related graph types.
Basic plot types
Boxplots
 |
Histograms
 |
Density plots
 |
Scatterplots
 |
Line graphs
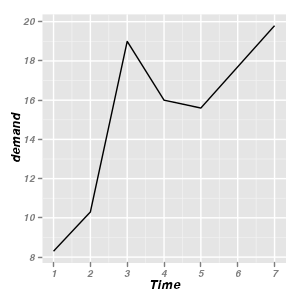 |
Smoothers
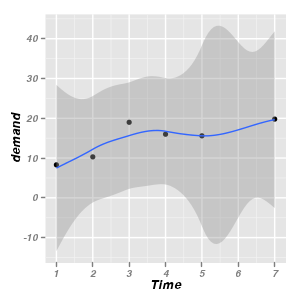 |
Trendlines
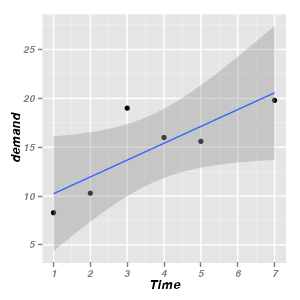 |
Bar charts
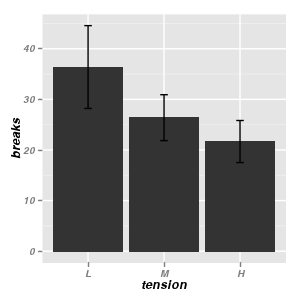 |
Stacked bar charts
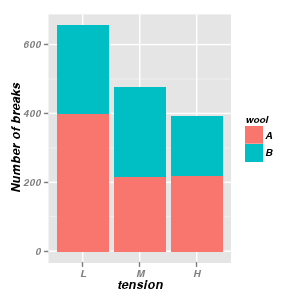 |
Bar graphcs
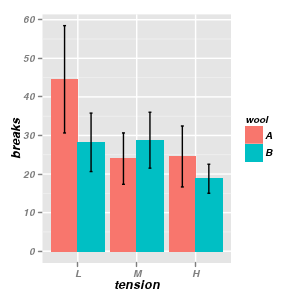 |
Interaction plots
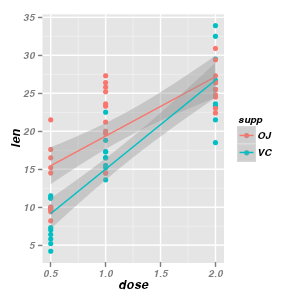 |
Scatterplot matrix
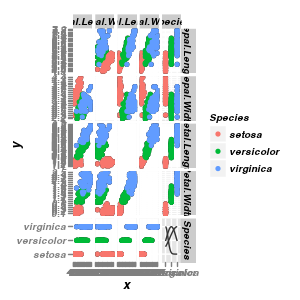 |
Heat maps
 |
Contour maps
 |
Additions
Segments
 |
Confidence bands
 |
Error bars
 |
Horizontal lines
 |
Vertical lines
 |
Range bars
 |
Axes rugs
 |
Text plots
 |
Customizations
Colours
 |
Line types
 |
Plotting symbols
 |
Transparency
 |
Sizes
 |
Date and time
 |
Layouts
facet_wrap
 |
facet_grid
 |
viewpots
 |
The Grammar of Graphics was first introduced/presented by Wilkinson
and Wills (2006) as a new graphics philosophy that laid down a series of
rules to govern
the production of quantitative graphics.
Essentially the proposed graphics infrastructure considers
a graphic as comprising a plot (defined by a coordinate
system, scales and panelling) over which one or more data
layers are applied.
Each layer is defined as:
- the data - a data frame
- mapping
specifications that
establish the visual aesthetics (colour, line type and
thickness, shapes etc) of each variable
- statistical methods that determine how the data rows
should be summarised (stat)
- geometric instructions
(geom) on how each
summary should be represented (bar, line, point etc)
- positional mechanism for dealing with
overlapping data (position)
The visual aspects of all the graphical features are then
governed by
themes.
Following a very short example, the next section will largely concentrate on describing each of the above graphical components.
Having then established the workings of these components, we can then put them together to yield specific graphics.
Hadley Wickham's interpretation of these principals in
an R context is implimented via the ggplot2
package. In addition the following packages are also commonly used alongside ggplot
so as to expand on the flexibility etc.
library(ggplot2)
library(grid)
library(gridExtra)
library(scales)
The following very simple graphic will be used to illustrate the above
specification by implicitly stating many of the default
specifications. It will use a cartesian coordinate system,
continuous axes scales, a single facet (panel) and then
define a single layer with a dataframe (BOD), with red
points, identity (no
summarisation) statistic visualised as a point geometric.
plotGgplot,
|
|
p <- ggplot() +
coord_cartesian() + #cartesian coordinates
scale_x_continuous() + #continuous x axis
scale_y_continuous() + #continuous y axis
#single layer
layer( data=BOD, #data.frame
mapping=aes(y=demand,x=Time),
stat="identity", #use original data
geom="point" #plot data as points
)+
layer( data=BOD, #data.frame
mapping=aes(y=demand,x=Time),
stat="identity", #use original data
geom="line" #plot data as a line
)
p #print the plot
OR, by leaving out all the default stuff
p <- ggplot(data = BOD, map = aes(y = demand, x = Time)) +
geom_point() + geom_line()
p
|
Note, the following important features of the grammar of graphics as implemented in R:
- the order in which each of the above components in the first code snippet were added is
unimportant. They each add additional information to the overall
graphical object. The object itself is evaluated as a whole when it is
printed.
- multiple layers are laid down in the order that they appear in the statement
- in the second code snippet (the shorter version), a layer is created for each of the two geoms
- the data and mapping used by both
geom_point() and geom_line are inherited from the main ggplot() function.
- since layers are ordered, the points are drawn first and the line over the top
In an attempt to illustrate the use of ggplot for elegant graphics,
we will drill down into each of the plot and layer specifications.
Although the geoms and thus layers are amongst the last features to be constructed by the system, the data and aesthetic features of the data
impact on how the coordinate system, scales and panelling work. Therefore, we will explore the geoms first.
Geometric objects - geom_ and stat_
Geometric objects (
geoms) are visual representations of observations. For example, there is a geom to represent points based on a set of x,y coordinates.
All graphics need at least one geom and each geom is mapped to
its own layer. Multiple geoms can be added to a graphic and the order that they are added to the expression determines the order that their respective
layer is constructed.
When a ggplot expression is being evaluated, geoms are coupled together with a stat_ function. This function is responsible
for generating data appropriate for the geom. For example, the stat_boxplot is responsible for generating the quantiles, whiskers
and outliers for the geom_boxplot function.
In addition to certain specific stat_ functions, all geoms can be coupled to a stat_identity function.
In mathematical contexts, identity functions map each element to themselves - this essentially means that each element passes straight through
the identity function unaltered. Coupling a geom to an identity function is useful when the characteristics of the data that you wish to
represent are present in the data frame. For example, your dataframe may contain the x,y coordinates for a series of points and you
wish for them to be used unaltered as the x,y coordinates on the graph. Moreover, your dataframe may contain pre-calculated information
about the quantiles, whiskers and outliers and you wish these to be used in the construction of a boxplot (rather than have the internals of ggplot perform the calculations on raw data).
Since geom_ and stats_ functions are coupled together, a geometric representation can be expressed from either
a geom_ function OR a stats_ function. That is, you either:
- specify a geom_ function that itself calls a stat_ function to provide the data for the geom function..
ggplot(CO2) + geom_smooth(aes(x = conc, y = uptake),
stat = "smooth")
- specify a stat_ function that itself calls a geom_ function to visually represent the data..
ggplot(CO2) + stat_smooth(aes(x = conc, y = uptake),
geom = "smooth")
It does not really make any difference which way around you do this. For the remainder of this tutorial, we will directly engage the
geom_
function for all examples.
The geom_ functions all have numerous arguments, many of which are common to all geoms_.
- data - the data frame containing the data. Typically this is inherited from the ggplot function.
- mapping - the aesthetic mapping instructions. Through the aesthetic mapping the aesthetic visual characteristics of the geometric features
can be controlled (such as colour, point sizes, shapes etc). The aesthetic mapping can be inherited from the ggplot function.
Common aesthetic features (mapped via a aes function) include:
- alpha - transparency
- colour - colour of the geometric features
- fill - fill colour of geometric features
- linetype - type of lines used in geometric features (dotted, dashed, etc)
- size - size of geometric features such as points or text
- shape - shape of geometric features such as points
- weight - weightings of values
- stat - the stat_ function coupled to the geom_ function
- position - the position adjustment for overlapping objects
- identity - leave objects were they are
- dodge - shift objects to the side to prevent overlapping
- stack - stack objects on top of each other
- fill - stack objects on top of each other and standardize each group to equal height
Currently, there are a large number of available geoms_ and stat_ functions within the ggplot system.
This tutorial is still a work in progress and therefore does not include all of them - I have focused on the more commonly used ones.
In an attempt to break up the set of geoms_ and stat_ functions, I have somewhat arbitrarily divided them up into
primary and secondary geometric features. Primary geometric features are those that could be viewed as graphics in their own right, whereas
secondary geometric features are those that are added to other geometric features to provide additional information (but would rarely be
considered a graphic in their own right).
Primary geometric objects
The following icon matrix provides navigation and an overview to the geometric features described in this section.
geom_bar
 |
geom_bar
 |
geom_bar
 |
geom_bar
 |
geom_boxplot
 |
geom_density
 |
geom_point
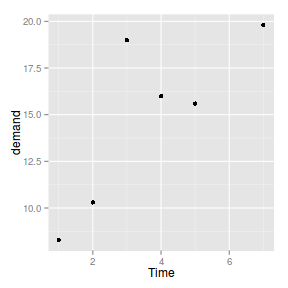 |
geom_line
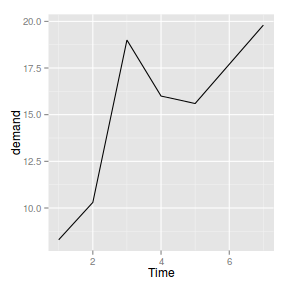 |
geom_smooth
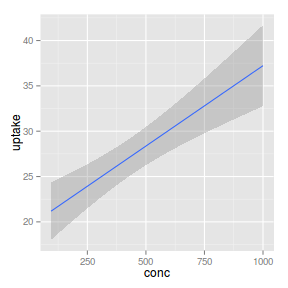 |
geom_smooth
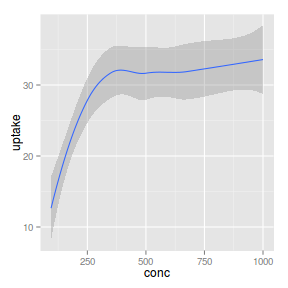 |
geom_tile
 |
geom_contour
 |
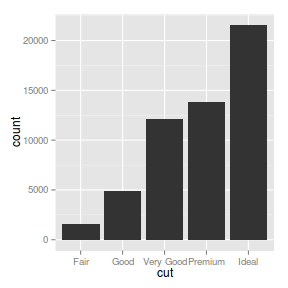
geom_bar and stats_bin
geom_bar constructs barcharts and histograms. By default, the bins of each bar along with the associated bar heights
are calculated by the
stats_bin function.
The following list describes the mapping aesthetic properties
associated with
geom_bar and
stats_bin. The entries in bold are compulsory.
| geom_bar | stat_bar |
- x - x axis value (categorical)
- alpha - transparency
- colour - colour of the lines
- fill - colour of the bar
- linetype - type of lines used to construct bar
- size - symbol size for outliers
- weight - weightings of values
|
- x - a vector that is to be binned
- y - optional y axis value (continuous)
|
The following table illustrates the first six rows of the
diamonds dataset (comes with R) that will be used for the following examples.
| | carat | cut | color | clarity | depth | table | price | x | y | z |
| 1 | 0.23 | Ideal | E | SI2 | 61.50 | 55.00 | 326 | 3.95 | 3.98 | 2.43 |
| 2 | 0.21 | Premium | E | SI1 | 59.80 | 61.00 | 326 | 3.89 | 3.84 | 2.31 |
| 3 | 0.23 | Good | E | VS1 | 56.90 | 65.00 | 327 | 4.05 | 4.07 | 2.31 |
| 4 | 0.29 | Premium | I | VS2 | 62.40 | 58.00 | 334 | 4.20 | 4.23 | 2.63 |
| 5 | 0.31 | Good | J | SI2 | 63.30 | 58.00 | 335 | 4.34 | 4.35 | 2.75 |
| 6 | 0.24 | Very Good | J | VVS2 | 62.80 | 57.00 | 336 | 3.94 | 3.96 | 2.48 |
| Feature | geom | stat | position | Aesthetic parameters / Notes | Example |
| barchart |
_bar |
_bin |
stack |
x,y,size,linetype,colour,fill,alpha, weight
bar heights represent number of items in each level of the categorical vector
|
|
ggplot(diamonds) + geom_bar(aes(x = cut))
|
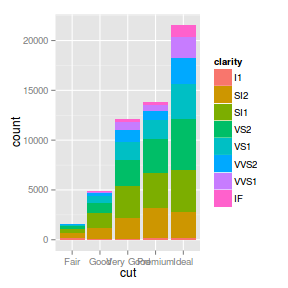
| barchart |
_bar |
_bin |
stack |
Bar heights represent the number of items in each level of a categorical vector and stacked
according to another categorical vector
|
|
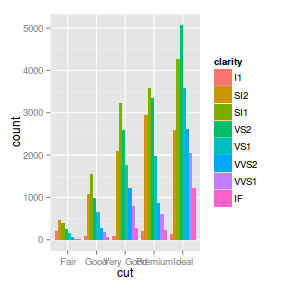
ggplot(diamonds) + geom_bar(aes(x = cut,
fill = clarity))
|
| barchart |
_bar |
_bin |
dodge |
bar heights represent the number of items in each combination of levels of multiple categorical vectors displayed side by side
|
|
ggplot(diamonds) + geom_bar(aes(x = cut,
fill = clarity), position = "dodge")
|
| barchart |
_bar |
_identity |
stack |
bar heights represent value of y for each x
|
|
diamonds1 <- as.data.frame(table(diamonds$cut))
ggplot(diamonds1) + geom_bar(aes(x = Var1,
y = Freq), stat = "identity")
|
| bargraph |
_bar |
_summary |
stack |
bar heights represent mean y within each level of categorical vector
|
|
ggplot(diamonds) + geom_bar(aes(x = cut,
y = carat), stat = "summary",
fun.y = mean)
|
| histogram |
_bar |
_bin |
stack |
bar heights represent counts within a binned continuous vector
|
|
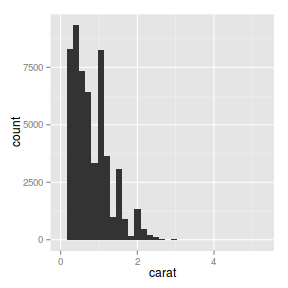
ggplot(diamonds) + geom_bar(aes(x = carat))
|
id="boxplot"geom_boxplot and stat_boxplot
geom_boxplot constructs boxplots. The values of the various elements of the boxplot (quantiles, whiskers etc)
are calculated by its main pairing function (
stat_boxplot). The following list describes the mapping aesthetic properties
associated with
geom_boxplot. The entries in bold are compulsory.
Note that boxplots are usually specified via the
geom_boxplot function which will engage the
stat_boxplot
to calculate the quantiles, whiskers and outliers. Therefore, confusingly, when calling
geom_boxplot, the compulsory paramters
are actually those required by
stat_boxplot (unless you indicated to use
stat_identity).
| geom_boxplot | stat_boxplot |
- x - x axis value (categorical)
- lower - value of the lower box line (25% percentile)
- middle - value of the middle box line (50% percentile - median)
- lower - value of the upper box line (75% percentile)
- ymax - value of the upper whisker
- ymax - value of the lower whisker
- alpha - transparency
- colour - colour of the lines
- fill - colour of the boxplot
- linetype - type of lines used to construct boxplot
- shape - symbol shape for outliers
- size - symbol size for outliers
- weight - weightings of values
|
- x - x axis value (categorical)
- y - y axis value (continuous)
|
| Feature | geom | stat | position | Notes, additional parameters | Example |
| boxplot |
_boxplot |
_boxplot |
dodge |
Plot of quantiles, whiskers and outliers
- outlier.colour
- outlier.shape
- outlier.size
- notch - whether to include a notch or not
- notchwidth - width of notch (fraction of box width)
|
|
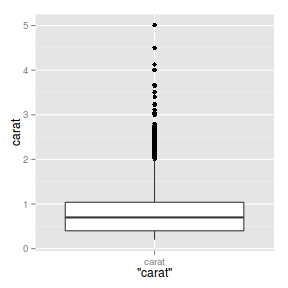
ggplot(diamonds) + geom_boxplot(aes(x = "carat",
y = carat))
|
geom_density and stat_density
geom_density constructs smooth density distributions from continuous vectors. The actual smoothed densities are
calculated by its main pairing function (
stat_density). The following list describes the mapping aesthetic properties
associated with
geom_density and
stat_density. The entries in bold are compulsory.
Note that density plots are usually specified via the
geom_density function which will engage the
stat_density.
Therefore, confusingly, when calling
geom_density, the compulsory paramaters
are actually those required by
stat_density (unless you indicated to use
stat_identity).
| geom_density | stat_density |
- x - x axis value (continuous)
- y - y axis value (densities)
- alpha - transparency
- colour - colour of the lines
- fill - colour of the density
- linetype - type of lines used to construct density
- shape - symbol shape for outliers
- size - symbol size for outliers
- weight - weightings of values
|
- x - a continuous vector from which to create density distribution
- fill - fill colour
- y
|
| Feature | geom | stat | position | Notes, additional parameters | Example |
| density |
_density |
_density |
dodge |
Density plot of a distribution of a vector
- adjust - smoothness
- kernel - kernel density
- trim - whether to trim densities to data range
|
|
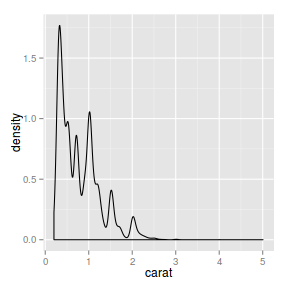
ggplot(diamonds) + geom_density(aes(x = carat))
|
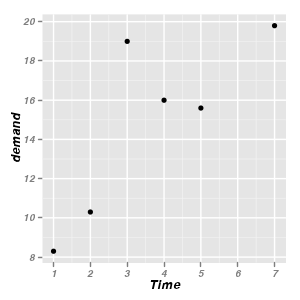
geom_point
geom_point draws points (scatterplot). Typically the stat used is
stat_identity as we wish to use the values
in two continuous vectors as the coordinates of each point. The following list describes the mapping aesthetic properties
associated with
geom_point. The entries in bold are compulsory.
| geom_point |
- x - x axis value (continuous)
- y - y axis value (densities)
- alpha - transparency
- colour - colour of the lines
- fill - colour of the point
- linetype - type of lines used to construct point
- shape - symbol shape for outliers
- size - symbol size for outliers
|
Note, it is possible to combine
geom_point with other stats (such as
stat_summary), so as to
plot summaries of the data rather than raw data.
| Feature | geom | stat | position | Notes, additional parameters | Example |
| point |
_point |
_identity |
identity |
Scatterplot
|
|
ggplot(BOD) + geom_point(aes(x = Time,
y = demand))
|
| means point |
_point |
_summary |
identity |
Means plot
|
|
ggplot(CO2) + geom_point(aes(x = conc,
y = uptake), stat = "summary",
fun.y = mean)
|
geom_line
geom_line draws lines joining coordinates. Typically the stat used is
stat_identity as we wish to use the values
in two continuous vectors as the coordinates of each line segment. The following list describes the mapping aesthetic properties
associated with
geom_line. The entries in bold are compulsory.
| geom_line |
- x - x axis value (continuous)
- y - y axis value (densities)
- alpha - transparency
- colour - colour of the lines
- fill - colour of the line
- linetype - type of lines used to construct line
- size - symbol size for outliers
|
Note, it is possible to combine
geom_line with other stats (such as
stat_summary), so as to
plot summaries of the data rather than raw data.
| Feature | geom | stat | position | Notes, additional parameters | Example |
| line |
_line |
_identity |
identity |
Line plot
|
|
ggplot(BOD) + geom_line(aes(x = Time,
y = demand))
|
| means line |
_line |
_summary |
identity |
Means line plot
|
|
ggplot(CO2) + geom_line(aes(x = conc,
y = uptake), stat = "summary",
fun.y = mean)
|
geom_smooth and stat_smooth
geom_smooth draws smooths lines (and 95% confidence intervals) through data clouds. Typically the stat used is
stat_smooth which in turn
engages one of the available smoothing methods (e.g. lm, glm, gam, loess or rlm).
The following list describes the mapping aesthetic properties
associated with
geom_smooth and
stat_smooth. The entries in bold are compulsory.
| geom_smooth | stat_smooth |
- x - x axis value (continuous)
- y - y axis value (densities)
- alpha - transparency
- colour - colour of the smooths
- fill - colour of the smooth
- linetype - type of smooths used to construct smooth
- size - symbol size for outliers
- weight - for weighting data
|
- x - x axis value (continuous)
- y - y axis value (densities)
|
stat_smooth also has the following optional arguments:
- method - the smoothing method (function). One of "lm", "glm", "gam", "loess" or "rlm"
- formula - the formula for the smoothing function, expressed relative to x and y. E.g. "y~x", "y~s(x)"
- se - whether to display confidence intervals
- fullrange - whether the fit should span the full range of the data
- level - confidence level (e.g. 0.95)
- n - number of points to evaluate smoother at
| Feature | geom | stat | position | Notes, additional parameters | Example |
| smooth |
_smooth |
_identity |
identity |
Linear smoother
|
|
ggplot(CO2) + geom_smooth(aes(x = conc,
y = uptake), method = "lm")
|
| Lowess smoother |
_smooth |
_stat |
identity |
Lowess smoother
|
|
ggplot(CO2) + geom_smooth(aes(x = conc,
y = uptake), method = "loess")
|
| Gam smoother |
_smooth |
_stat |
identity |
Cupic regression spline smoother
|
|
library(mgcv)
ggplot(CO2) + geom_smooth(aes(x = conc,
y = uptake), method = "gam",
formula = y ~ s(x, bs = "cr",
k = 4))
|
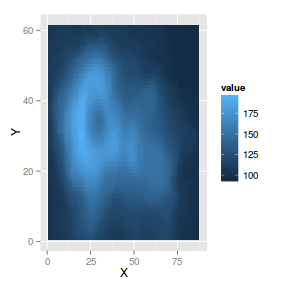
geom_tile
geom_tile constructs heat maps given x,y coordinates and a z value to associate with the fill of each tile.
The following list describes the mapping aesthetic properties
associated with
geom_tile and
stat_tile. The entries in bold are compulsory.
| geom_tile |
- x - x axis value (continuous)
- y - y axis value (continuous)
- alpha - transparency
- colour - colour of the borders around tiles
- fill - colour of the fill of tiles
- linetype - type of lines to use as borders to each tile
- size - line thickness
|
| Feature | geom | stat | position | Notes, additional parameters | Example |
| tile |
_identity |
_identity |
identity |
Heat map
|
|
library(reshape)
volcano.df <- melt(volcano, varnames = c("X",
"Y"))
ggplot(volcano.df) + geom_tile(aes(x = X,
y = Y, fill = value))
|
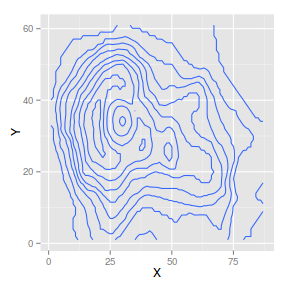
geom_contour and stat_contour
geom_contour constructs contour maps given x,y coordinates and a z value from which to calculate each contour.
The following list describes the mapping aesthetic properties
associated with
geom_contour and
stat_contour. The entries in bold are compulsory.
| geom_contour | stat_contour |
- x - x axis value (continuous)
- y - y axis value (continuous)
- alpha - transparency
- colour - colour of the contour lines
- linetype - line type of the contour lines
- size - line thickness
- weight
|
- x - x axis value (continuous)
- y - y axis value (continuous)
- z - z axis value (continuous)
- order
|
| Feature | geom | stat | position | Notes, additional parameters | Example |
| contour |
_identity |
_identity |
identity |
Heat map
|
|
library(reshape)
volcano.df <- melt(volcano, varnames = c("X",
"Y"))
ggplot(volcano.df) + geom_contour(aes(x = X,
y = Y, z = value))
|
Secondary geometric objects
geom_segment
 |
geom_ribbon
 |
geom_errorbar
 |
geom_hline
 |
geom_vline
 |
geom_pointrange
 |
geom_rug
 |
geom_text
 |
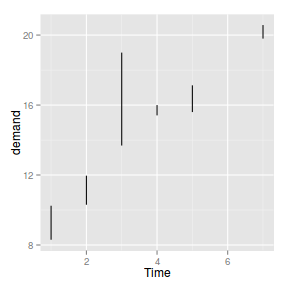
geom_segment
geom_segment draws segments joining coordinates. The following list describes the mapping aesthetic properties
associated with
geom_segment. The entries in bold are compulsory.
| geom_segment |
- x - x coordinates for the start of lines
- xend - x coordinates for the end of lines
- y - y coordinates for the start of lines
- yend - y coordinates for the end of lines
- alpha - transparency
- colour - colour of the segments
- fill - colour of the segment
- linetype - type of segments used to construct segment
- size - symbol size for outliers
|
geom_segment also has the following optional arguments:
- arrow - specification of how arrows should be constructed
- lineend - style of the line end
| Feature | geom | stat | position | Notes, additional parameters | Example |
| segment |
_identity |
_identity |
identity |
Segments on a plot - useful for drawing lots of lines or arrows
|
|
BOD.lm <- lm(demand ~ Time, data = BOD)
BOD$fitted <- fitted(BOD.lm)
BOD$resid <- resid(BOD.lm)
ggplot(BOD) + geom_segment(aes(x = Time,
y = demand, xend = Time, yend = fitted))
|
| segment |
_identity |
_identity |
identity |
Segments on a plot - useful for drawing lots of lines or arrows
|
|
BOD.lm <- lm(demand ~ Time, data = BOD)
BOD$fitted <- fitted(BOD.lm)
BOD$resid <- resid(BOD.lm)
ggplot(BOD) + geom_segment(aes(x = Time,
y = demand, xend = Time, yend = fitted),
arrow = arrow(length = unit(0.5,
"cm")))
|
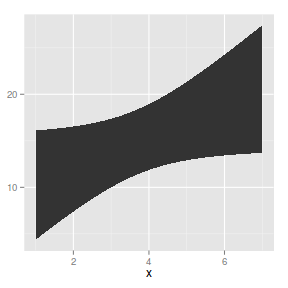
geom_ribbon
geom_ribbon draws ribbons based on upper and lower levels of y associated with each level of x.
The following list describes the mapping aesthetic properties
associated with
geom_ribbon. The entries in bold are compulsory.
| geom_ribbon |
- x - x coordinates
- ymin - y coordinates of the lower limits
- ymax - y coordinates of the upper limits
- alpha - transparency
- colour - colour of the ribbons
- fill - colour of the ribbon
- linetype - type of lines used to construct the borders of the ribbon
- size - thickness of the lines used to border the ribbon
|
| Feature | geom | stat | position | Notes, additional parameters | Example |
| ribbon |
_identity |
_identity |
identity |
Ribbons on a plot - useful for depicting confidence envelopes
|
|
BOD.lm <- lm(demand ~ Time, data = BOD)
xs <- seq(min(BOD$Time), max(BOD$Time),
l = 100)
pred <- data.frame(predict(BOD.lm,
newdata = data.frame(Time = xs),
interval = "confidence"))
pred$x <- xs
ggplot(pred) + geom_ribbon(aes(x = x,
ymin = lwr, ymax = upr))
|
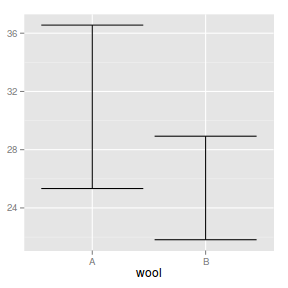
geom_errorbar
geom_errorbar draws errorbars based on upper and lower levels of y associated with each level of x.
The following list describes the mapping aesthetic properties
associated with
geom_errorbar. The entries in bold are compulsory.
| geom_errorbar |
- x - x coordinates
- ymin - y coordinates of the lower limits
- ymax - y coordinates of the upper limits
- alpha - transparency
- colour - colour of the errorbars
- fill - colour of the errorbar
- linetype - type of lines used to construct the borders of the errorbar
- size - thickness of the lines used to border the errorbar
- width - width of the errorbars
|
| Feature | geom | stat | position | Notes, additional parameters | Example |
| errorbar |
_identity |
_identity |
identity |
Error bars on a plot - useful for adding to means plots etc
|
|
library(plyr)
warpbreaks.df <- ddply(warpbreaks,
~wool, function(x) {
Hmisc:::smean.cl.boot(x$breaks)
})
ggplot(warpbreaks.df) + geom_errorbar(aes(x = wool,
ymin = Lower, ymax = Upper))
|
| errorbar |
_identity |
_summary |
identity |
Error bars on a plot - useful for adding to means plots etc
|
|
ggplot(warpbreaks) + geom_errorbar(aes(x = wool,
y = breaks), stat = "summary",
fun.data = "mean_cl_boot")
|
library(ggmap)
hdf <- get_map(location=c(lon=146.8,lat=-19.20), zoom=12, maptype="satellite")
ggmap(hdf, extent = 'normal')
Coordinate system - coord
The coordinate system controls the nature and scale of the axes.
| System | Parameters | Example |
Regular cartesian coordinate system
coord_cartesian |
xlim - x limits
ylim - y limits |
|
ggplot(BOD) + coord_cartesian() +
geom_line(aes(y = demand, x = Time))
|
Polar coordinate system
coord_polar |
theta="x" - angle variable
start=0 - initial angle from 12 oclock
direction=1 - 1=clockwise, -1=anticlockwise |
|
ggplot(BOD) + coord_polar() + geom_line(aes(y = demand,
x = Time))
|
Flipped the axes
coord_flip |
xlim=NULL - y limits
ylim=NULL - x limits |
|
ggplot(BOD) + coord_flip() + geom_line(aes(y = demand,
x = Time))
|
Fix the ratio of axes dimesions
coord_fixed |
ratio=1 - y/x ratio
xlim=NULL - x limits
ylim=NULL - y limits |
|
ggplot(BOD) + coord_fixed(ratio = 0.25) +
geom_line(aes(y = demand, x = Time))
|
1:1 (equal) ratio of axes dimesions
same as coord_fixed(ratio=1)
coord_equal |
xlim=NULL - x limits
ylim=NULL - y limits |
|
ggplot(BOD) + coord_equal() + geom_line(aes(y = demand,
x = Time))
|
Map projection coordinate system
coord_map |
projection="mercator" - mapping projection
orientation=c(90,0,mean(range(x))) - map orientation |
|
# get high resolution map of
# Australia (and islands) data
library(maps)
library(mapdata)
aus <- map_data("worldHires", region = "Australia")
# Orthographic coordinates
ggplot(aus, aes(x = long, y = lat,
group = group)) + coord_map("ortho",
orientation = c(-20, 125, 23.5)) +
geom_polygon()
|
Altering the axes scales via the coordinate system
Modifying scales with coords affects the zoom on the graph.
That is, it defines the extent and nature of the axes coordinates.
By contrast, altering limits via scale_ routines will alter the scope of data
included in a manner analogous to operating on a subset of the data.
| Default scale | Scale via coord_ (Zoom) | Scale via scale_ |
|
|
|
|
# Default scales
ggplot(BOD, aes(y = demand, x = Time)) + geom_point() + geom_smooth(method = "lm")
# Zoom on x-axis
ggplot(BOD, aes(y = demand, x = Time)) + coord_cartesian(xlim = c(2, 6)) + geom_point() + geom_smooth(method = "lm")
# Scale (subset) the x data
ggplot(BOD, aes(y = demand, x = Time)) + scale_x_continuous(limits = c(2, 6)) + geom_point() + geom_smooth(method = "lm")
|
In addition to altering the zoom of the axes, axes (coordinate system) scales can be transformed to other scales via the coord_trans
function. Transformations of the coordinate system take place after statistics have been calculated and geoms derived.
Therefore the shape of geoms are altered.
The coord_trans() function has the following argments:
- xtrans
- a transformer that will operate on the x scale
- ytrans
- a transformer that will operate on the y scale
- limx
- limits of the x axis
- limy
- limits of the y axis
A transformer is a function that defines a transformation along with its inverse and rules on how to generate pretty breaks (tick marks) and their labels
To illustrate the distinction between
coord_trans and
scale_, we will generate some curvilinear data.
set.seed(1)
n <- 50
dat <- data.frame(x = exp((1:n + rnorm(n, sd = 2))/10), y = 1:n + rnorm(n, sd = 2))
| Linear scales | coord_trans | scales_ |
|
Linear spacing of axis ticks
|
Log10 spacing of axis ticks on a linear scale
|
Linear spacing of axis ticks on a log10 scale
|
ggplot(dat, aes(y = y, x = x)) + geom_point()
ggplot(dat, aes(y = y, x = x)) + geom_point() + coord_trans(xtrans = log10_trans())
ggplot(dat, aes(y = y, x = x)) + geom_point() + scale_x_continuous(trans = log10_trans())
|
|
Linear trend applied to curved data
|
Linear trend applied to curved data, then bent by coordinates rescaling
|
Linear trend applied to scaled (linear) data
|
ggplot(dat, aes(y = y, x = x)) + geom_point() + geom_smooth(method = "lm")
ggplot(dat, aes(y = y, x = x)) + geom_point() + geom_smooth(method = "lm") + coord_trans(xtrans = log10_trans())
ggplot(dat, aes(y = y, x = x)) + geom_point() + geom_smooth(method = "lm") + scale_x_continuous(trans = log10_trans())
|
Transformers
trans_new
The trans_new function itself defines and returns a list structure comprising;
- name
A name to be given to the transformation
- transform
a function (or name of a function) that performs the transformation
- inverse
a function (or name of a function) that performs the inverse of the transformation
- breaks
a function that generates tick breaks. Operates on the raw data
- format
a function that formats labels for the breaks.
- domain
the range over which the transformation is valid
To illustrate the trans_new function, lets define a natural log (ln) transformer to apply to our artificial data.
ggplot(dat, aes(y=y,x=x)) + geom_point() +
geom_smooth(method="lm") +
scale_x_continuous(trans=trans_new(name="ln",
transform=function(x) log(x),
inverse=function(x) exp(x),
breaks=function(x) pretty(x),
domain=c(1e-100,Inf)))
|
## Error: 'from' cannot be NA, NaN or infinite
|
ln_trans <- function() {
name <- "ln"
trans <- function(x) log(x)
inv <- function(x) exp(x)
breaks <- function(x) pretty(x)
format <- function(x) x
domain <- c(1e-100,Inf)
trans_new(name,transform=trans,inverse=inv,
breaks=breaks, domain=domain)
}
ggplot(dat, aes(y=y,x=x)) + geom_point()+
geom_smooth(method="lm")+
scale_x_continuous(trans=ln_trans())
|
## Error: 'from' cannot be NA, NaN or infinite
|
coord_trans(xtrans = "identity", ytrans = "identity",limx = NULL, limy = NULL)
Finer control of the transformation can be exersized. Consider the following examples using the same dataset.
In order to be able to use trans_new effectively, it is necessary to understand the data parsed to each of the functions within the transformer.
In the following demonstration, I have placed print statements within each of the functions so as to illustrate the sequence in which the functions are called (relative to each other and other external functions) as well
as the input data of each function. Note for this demonstration, I have ommitted the smoother as it would also result in calls to these functions and therefore
compound the sequencing.
p<-ggplot(dat, aes(y=y,x=x)) + geom_point()+# + geom_smooth(method="lm") +
scale_x_continuous(trans=trans_new(name="",transform=function(x) {
cat("**Tranform begin**\n");
print(x);
log10(x);
},
inverse=function(x) {
cat("**Inverse**\n");
print(x);
10^(x);
},
breaks=function(x) {
cat("**Breaks**\n");
print(x);
pretty(x);
},
format=function(x) {cat("**Format**\n");print(x);x;},
domain=c(1e-100,Inf)))
p+ theme(plot.background = element_rect(fill = "transparent",colour = NA))
**Tranform begin**
[1] 0.975 1.267 1.142 2.052 1.761
[6] 1.546 2.220 2.580 2.760 2.557
[11] 4.065 3.589 3.241 2.604 5.612
[16] 4.909 5.456 7.307 7.879 8.321
[21] 9.814 10.553 10.124 7.405 13.790
[26] 13.313 14.423 12.254 16.517 21.837
[31] 29.129 24.033 29.298 29.643 25.143
[36] 33.683 37.380 44.174 61.560 63.601
[41] 58.387 63.391 84.723 91.043 78.433
[46] 86.358 118.264 141.699 131.306 177.013
**Inverse**
[1] -0.1239 2.3610
**Breaks**
[1] 0.7517 229.5905
**Tranform begin**
[1] 0.7517 229.5905
**Tranform begin**
[1] 0 50 100 150 200 250
**Inverse**
[1] -Inf 1.699 2.000 2.176 2.301 NA
**Format**
[1] 0 50 100 150 200 NA
Error: 'from' cannot be NA, NaN or infinite
The sequence is as follows;
- The first call to the **Transform** function is parsed the raw data
- The first call to the **Inverse** function is parsed the computed axes limits (on the log10 scale).
These originate in another part of the ggplot engine.
Following the action of the transformation function, other functions determine the limits of the axes based on the transformed data as well as the nominated axis expansion factor
(places a buffers beyond the data such that geoms do not overlapp axes).
The inverse function then converts these limits into limits in the original raw data scale.
- The first call to the **Breaks** function is parsed the axes limits on the scale of the raw data and defines the spacing of axes tick marks
- The second call to the **Transform** function takes the axis limits and rescales them into the log10 scale
- The third call to the **Transform** function takes the axis tick mark spacing from **Breaks** and rescales them into the log10 scale
- The second call to the **Inverse** function takes the axis tick marks spacing on the log10 scale and rescales into the scale of the raw data
- Finally, the **Format** function is used to define the labels to be applied to the tick marks
| Axes in the scale of observations |
|
|
|
|
|
|
Axes in the scale of logarithms |
|
|
|
|
|
|
*_trans transformers
The _trans family of transformers are convienient wrappers for the trans_new function.
| Transformer | Desciption |
| asn_trans() |
Arc-sin square-root transformation (of proportions/percentages). |
| atanh_trans() |
Arc-tangent transformation |
| boxcox_trans(p) |
Box-Cox power transformation
When the power exponent (p) is equal to 0, values are logged
For exponents other than zero, 1 is subtracted from the value are raised to the power of the exponent and this is then divided by the exponent. |
| date_trans |
| exp_trans |
| identity_trans |
| log10_trans |
| log1p_trans |
| log2_trans |
| log_trans |
| logit_trans |
| probability_trans |
| probit_trans |
| reciprocal_trans |
| reverse_trans |
| sqrt_trans |
| time_trans |
| Transform axes scale (logs) | 1:1 axes scales |
|
|
# log10 axes scales
ggplot(BOD) + coord_trans(xtrans = "log10", ytrans = "log10") +
geom_line(aes(y = demand, x = Time))
|
Modifying scales with coords affects the zoom on the graph.
That is, it defines the extent and nature of the axes coordinates.
By contrast, altering limits via scale_ routines will alter the scope of data
included in a manner analogous to operating on a subset of the data.
Scales
The idea of scales is that you present the plotting engine with data
or characteristics in one scale and use the various
scale_ functions to convert those data into
another scale. In the grammar of graphics, scales are synonymous for units of data, colors, shapes, sizes etc of plotting features
and the axes and guides (legends) provide a visual cue for what the scales are. For example;
- you might include data that ranges from
10 to 20 units, yet you wish to produce a plot that zooms in on the
range 12-16.
- you have presented grouped data (data with multiple
trends) and instructed the graphing engine to assign different
colour codes to each trend. You can then
define a colour scale to adjust the exact colours rendered.
- similarly, you might have indicated how plotting symbol shape
and size are to be distinguished in your data set. You can then
assign scales that define the exact shapes and symbol sizes rendered.
Technically, scales determine how attributes of the data are mapped into aesthetic geom properties.
The majority of geom's (geometric objects) have the following aesthetic properties:
- x - the x position (coordinates) of the geom
- y - the y position (coordinates) of the geom
- size - the size of the geom (e.g. the size of a point)
- shape - the shape of the geom
- linetype - the type of line associated with the geom's outline (solid, dashed etc)
- colour - the colour of the geom's outline (note the English spelling of the word colour)
- fill - the colour of the geom's fill
- alpha - the transparency of the geom (0=transparent, through to 1=opaque)
In turn, each of these properties are mapped to a scale - the defaults of which are automatically selected
according to what is appropriate for the sort of data. For example, data can be on a continuous or discrete (categorical)
scale. Most data type have the following possible scales for each of the above properties:
_continuous - when you want the scale increments (such as the different point sizes, colours etc) to be determined from a continuous vector in your data frame._discrete - when you want the scale increments (such as the different point sizes colours etc) to be determined from a categorical vector in your data frame._manual - is a variation on _discrete and is used when you wish to manually indicate the characteristic of each increment. You need to provide as many
values as there are levels of your discrete vector._identity - is another variation on _discrete and is used when you wish for the values in your categorical vector
to be used un-scaled as the characteristics of the data. For example, your data frame might contain a vector of colour names or point sizes.
Some properties, such as colour also have additional scales that are specific to the characteristic.
The scales effect not only the characteristics of the geoms, they also effect the guides (legends) that accompany the geoms.
Scaling functions comprise the prefix scale_, followed by the name of an aesthetic property and suffixed by the type of scale.
Hence a function to manually define a colour scale would be scale_colour_manual.
All scales have the following arguments available:
name - a title applied to the scale. In the case of scales for x and y (the x,y coordinates of geoms), the name is the axis title.
For all other scales, the name is the title of the guide (legend).breaks - the increments on the guide. For scale_x_ and scale_y_, breaks are the axis tick locations.
For all other scales, the breaks indicate the increments of the characteristic in the legend (e.g. how many point shapes are featured in the legend).labels - the labels given to the increments on the guide. For scale_x_ and scale_y_, labels are the axis tick labels.
For all other scales, the labels are the labels given to items in the legend.limits - the span/range of data represented in the scale. Note, if the range is inside the range of the data, the data are sub-setted.trans - scale transformations applied - obviously this is only relevant to scales that are associated with continuous data.
Scaling the x and y values (scale_x_)
The
scale_x_ and
scale_y_ scales control the x and y axes and in addition to the common arguments listed above,
the following optional arguments available for specific scales:
expand - a vector of length two that indicates multiplicative and additive constants used to expand the axes away from the data thereby ensuring that geoms do not intersect with the axes.minor_breaks - the increments for the minor breaks along the axis. The minor breaks have a grid line yet no tick marks or labels.
scale_x_continuous | scale_x_continuous | scale_x_continuous |
| linear scaling |
linear with nice title |
linear with more space |
|
|
|
|
# Linear axes scales with altered axis title
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
scale_x_continuous(name = "CO2 conc")
# Linear axes scales with more complex title
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
scale_x_continuous(name = expression(paste("Ambient ",
CO[2], " concentration (mg/l)", sep = "")))
# Linear axes scales with more space along the x
# axis
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
scale_x_continuous(name = "CO2 conc", expand = c(0,
200))
|
scale_x_log10 | scale_x_sqrt | scale_x_reverse |
| Log10 scale |
Square-root scale |
Reverse scale |
Shortcut for
scale_x_continuous(trans=
log10_trans())
|
Shortcut for
scale_x_continuous(trans=
sqrt_trans())
|
|
# log10 axes scales
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
scale_x_log10(name = "CO2 conc", breaks = as.vector(c(1,
2, 5, 10) %o% 10^(-1:2)))
# square-root transformation
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
scale_x_sqrt(name = "CO2 conc")
# reverse the data
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
scale_x_reverse(name = "CO2 conc")
|
scale_x_date | scale_x_datetime | scale_x_discrete |
For more info on date breaks see date_breaks
For more date formats see strptime
|
For more info on date breaks see date_breaks
For more date formats see strptime
|
|
# Date format
library(scales)
CO2$Date <- as.Date(paste(2000 + as.numeric(as.factor(CO2$conc)),
"-01-01", sep = ""))
ggplot(CO2, aes(y = uptake, x = Date)) + geom_point() +
scale_x_date(name = "Year", breaks = "2 years",
minor_breaks = "6 month", labels = date_format("%Y"))
# POSIX format
library(scales)
CO2$DateTime <- as.POSIXct(paste(2000, "-0", as.numeric(as.factor(CO2$conc)),
"-01 09:00:00", sep = ""))
ggplot(CO2, aes(y = uptake, x = DateTime)) + geom_point() +
scale_x_datetime(name = "Time (days)", breaks = "2 months",
minor_breaks = "1 months", labels = date_format("%b"))
# categorical axis
ggplot(CO2, aes(y = uptake, x = Treatment)) + geom_point() +
scale_x_discrete(name = "Treatment")
|
Scaling the size of geoms (scale_size_)
The
scale_size_ scales control the size of geoms (such as the size of points) and in addition to the common scale arguments, the following optional arguments available:
range - the minimum and maximum sizevalues - the specific sizes to use (for _manual scale)guide - whether to include a guide and what sort of guide to include (e.g. "legend")
scale_size_continuous | scale_size_discrete | scale_size_manual |
| Scale the geoms according to a continuous vector |
Scale the geoms according to a categorical vector |
Manually determine the size of geoms |
range - minimum and maximum geom size
|
|
values - a set of values to use for sizes
|
# size determined by continuous covariate
set.seed(123)
CO2$cv <- runif(nrow(CO2), 10, 50)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(size = cv)) +
scale_size_continuous(name = "Temperature")
# Discrete sizes ranging in size from 2 to 4
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(size = Type)) +
scale_size_discrete(name = "Type", range = c(2,
4))
# Manual sizes of exactly 2 and 4
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(size = Type)) +
scale_size_manual(name = "Type", values = c(2,
4))
|
scale_size_identity | | |
| Size the geoms according to the values of a continuous vector (don't scale) |
|
|
guide - whether to include a guide (legend)
|
# Sizes provided by a covariate
set.seed(123)
CO2$Count <- runif(nrow(CO2), 0, 10)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(size = Count)) +
scale_size_identity(name = "Type", guide = "legend")
|
Scaling the shape of geoms (scale_shape_)
The
scale_shape_ scales control the shape of geoms (such as the shape of the plotting point) an in addition to all of the regular arguments, the following optional arguments are available:
solid - whether the shapes should be solid (TRUE) or outlined (FALSE)
scale_shape_discrete | scale_shape_manual | scale_shape_identity |
| Geom shapes determined (scaled) by categorical variable |
Geom shapes determined (scaled) manually |
Geom shapes determined by categorical variable (no scaling) |
|
|
values - a set of values (or shape names) to use for shapes
|
|
# Discrete shapes determined by the combination of
# Type and Treatment The items in the guide are
# then rearranged and re-labelled
CO2$Comb <- interaction(CO2$Type, CO2$Treatment)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(shape = Comb)) +
scale_shape_discrete(name = "Type", breaks = c("Quebec.nonchilled",
"Quebec.chilled", "Mississippi.nonchilled",
"Mississippi.chilled"), labels = c("Quebec non-chilled",
"Quebec chilled", "Miss. non-chilled", "Miss. chilled"))
# Manual shapes
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(shape = Treatment),
size = 2) + scale_shape_manual(name = "Treatment",
values = c(16, 21))
# Identity shapes
set.seed(123)
CO2$Count <- runif(nrow(CO2), 0, 10)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(shape = Count)) +
scale_shape_identity(name = "Species", guide = "legend")
|
Scaling the linetype associated with geoms (scale_linetype_)
The
scale_size_ scales control the type of lines used in geoms and have the following additional optional arguments available:
values - values supplied to manually determine the line types
scale_linetype_discrete | scale_linetype_manual | scale_linetype_identity |
| Geom linetypes determined (scaled) by categorical variable |
Geom linetypes determined (scaled) manually |
Geom linetypes determined by categorical variable (no scaling) |
|
|
values - a set of values (or linetype names) to use for linetypes
|
|
# Discrete shapes determined by the combination of
# Type and Treatment The items in the guide are
# then rearranged and re-labelled
CO2$Comb <- interaction(CO2$Type, CO2$Treatment)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth(aes(linetype = Comb)) +
scale_linetype_discrete(name = "Type", breaks = c("Quebec.nonchilled",
"Quebec.chilled", "Mississippi.nonchilled",
"Mississippi.chilled"), labels = c("Quebec non-chilled",
"Quebec chilled", "Miss. non-chilled", "Miss. chilled"))
# Manual linetypes
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth(aes(linetype = Treatment)) +
scale_linetype_manual(name = "Treatment", values = c("dashed",
"dotted"))
# Identity linetypes
CO2$Lines <- factor(CO2$Treatment, levels = c("nonchilled",
"chilled"), labels = c("dotted", "dashed"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth(aes(linetype = Lines)) +
scale_linetype_identity(name = "Temperature", guide = "legend",
breaks = c("dotted", "dashed"), labels = c("Low",
"High"))
|
Scaling the colour (or fill) associated with geoms (scale_colour_ & scale_fill_)
The
scale_size_ scales control the colour of geoms and have the following additional optional arguments available:
low - colour of low end of the colour spectrumhigh - colour of high end of the colour spectrumguide - what sort of legend (e.g. colorbar)
scale_colour_continuous | scale_colour_gradient | scale_colour_gradient2 |
| Geom colours determined (scaled) by continuous variable |
Geom colours determined (scaled) palette |
Geom colours determined by a different palette |
|
|
|
|
# colour determined by continuous covariate
set.seed(123)
CO2$cv <- runif(nrow(CO2), 10, 50)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = cv)) +
scale_colour_continuous(name = "Temperature", low = "blue",
high = "red")
# colour determined by continuous covariate
set.seed(123)
CO2$cv <- runif(nrow(CO2), 10, 50)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = cv)) +
scale_colour_gradient(name = "Temperature")
# colour determined by continuous covariate
set.seed(123)
CO2$cv <- runif(nrow(CO2), 10, 50)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = cv)) +
scale_colour_gradient2(name = "Temperature")
|
scale_colour_gradientn | | |
| Geom colours determined (scaled) a specific palette |
|
|
|
|
# colour determined by continuous covariate use a
# predefined gradient based colour palette
set.seed(123)
CO2$cv <- runif(nrow(CO2), 10, 50)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = cv)) +
scale_colour_gradientn(name = "Temperature", colours = terrain.colors(5))
|
scale_colour_hue | scale_colour_grey | scale_colour_brewer |
| Evenly spaced geom colours determined (scaled) by hue |
Geom colours determined (scaled) palette |
Geom colours determined by a different palette |
|
|
|
See the color brewer site for more info
|
# Discrete colours for hue
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("Low", "Medium", "High"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = Temp)) +
scale_colour_hue(name = "Temperature", l = 80,
c = 130)
# Discrete colours
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("Low", "Medium", "High"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = Temp)) +
scale_colour_grey(name = "Temperature", start = 0.2,
end = 0.8)
# Discrete colours selected from a colour brewer
# palette it automatically knows how many colours
# are required
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("Low", "Medium", "High"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = Temp)) +
scale_colour_brewer(name = "Temperature", type = "seq",
palette = "Reds")
|
scale_colour_manual | scale_colour_identity | |
| Geom colours determined (scaled) a specific palette |
|
|
|
|
|
# Manual colours
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("Low", "Medium", "High"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(colour = Temp)) +
scale_colour_manual(name = "Temperature", values = c("red",
"#00AA00", 1))
# identity colours
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("red", "#00AA00", 1))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth(aes(colour = Temp)) +
scale_colour_identity(name = "Temperature", guide = "legend",
labels = c("Low", "Medium", "High"))
|
Scaling the alpha level of colour associated with geoms (scale_alpha_)
The
scale_alpha_ scales control the transparency of geoms and have the following additional optional arguments available:
range - the alpha range (0,1)values - alpha values between 0 and 1guide - what sort of legend (e.g. colorbar)
scale_alpha_continuous | scale_alpha_discrete | scale_alpha_manual |
| Evenly spaced geom alphas determined (scaled) by continuous |
Geom alphas determined (scaled) palette |
Geom alphas determined by a different palette |
|
|
|
|
# colour determined by continuous covariate
set.seed(123)
CO2$cv <- runif(nrow(CO2), 10, 50)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(alpha = cv)) +
scale_alpha_continuous(name = "Temperature", range = c(0.3,
1))
# Discrete alphas
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("Low", "Medium", "High"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(alpha = Temp)) +
scale_alpha_discrete(name = "Temperature")
# Manual alphas
set.seed(123)
CO2$cv <- runif(nrow(CO2), 0, 100)
CO2$Temp <- cut(CO2$cv, breaks = c(0, 33, 66, 100),
labels = c("Low", "Medium", "High"))
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(alpha = Temp)) +
scale_alpha_manual(name = "Temperature", values = c(0.3,
0.6, 0.95))
|
scale_alpha_identity | | |
| Geom alphas determined (scaled) a specific palette |
|
|
|
|
# Identity alphas
set.seed(123)
CO2$Alpha <- runif(nrow(CO2), 0, 1)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point(aes(alpha = Alpha)) +
scale_alpha_identity(name = "Temperature")
|
Facets (panels)
Faceting splits the data up into a matrix of panels on the basis of one or more categorical vectors. Since
facets display subsets of the data, they are very useful for examining trends in hierarchical designs.
There are two faceting function, that reflect two alternative approaches:
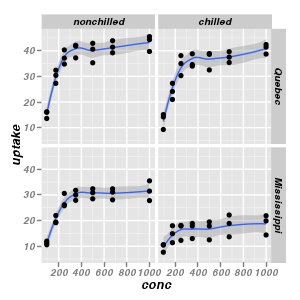
facet_wrap(~cell) - creates a set of panels based on a factor and wraps the panels into a 2-d matrix. cell
represents a categorical vector or set of categorical vectorsfacet_wrap(row~column) - creates a set of panels based on a factor and wraps the panels into a 2-d matrix.
row and column represents the categorical vectors used to define the rows and columns of the matrix respectively
facet_wrap
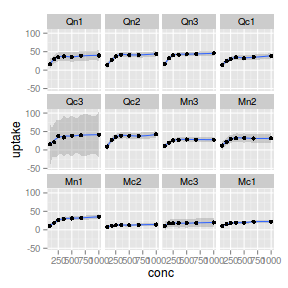 |
facet_grid
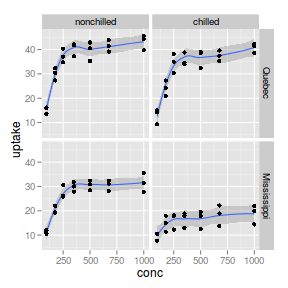 |
The following list describes the mapping aesthetic properties
associated with
facet_wrap and
facet_grid functions. The entries in bold are compulsory.
| facet_wrap | facet_grid |
- facets - formula specifying faceting variables to use in faceting
- nrow - number of rows
- ncol - number of columns
- scales - should all scaled be fixed or free
- "fixed" - (default) all scales the same
- "free" - all scales free
- "free_x" - all x-axis scales free
- "free_y" - all y-axis scales free
- as.table - if TRUE, laid out from top left to bottom right, if FALSE: bottom left to top right
- drop - drop factor combinations that lack data
|
- facets - formula specifying faceting variables to use in faceting
- margins - whether to include marginal trends
- scales - should all scaled be fixed or free
- "fixed" - (default) all scales the same
- "free" - all scales free
- "free_x" - all x-axis scales free
- "free_y" - all y-axis scales free
- space- should all panels take up the same space
- "fixed" - (default) all panels take up the same space
- "free" - panel heights and widths vary
- "free_x" - panel widths vary
- "free_y" - panel heights vary
- labeller - a function used to label the panel strips
- as.table - if TRUE, laid out from top left to bottom right, if FALSE: bottom left to top right
- drop - drop factor combinations that lack data
|
| Facet | Notes, additional parameters | Example |
_wrap |
Matrix of panels split by a single categorical vector
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + facet_wrap(~Plant)
|
_wrap |
Matrix of panels split by a single categorical vector with different y-axis scale range for each panel
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + facet_wrap(~Plant, scales = "free_y")
|
_grid |
Matrix of panels split by a single categorical vector with different y-axis scale range for each panel
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + facet_grid(Type ~ Treatment)
|
_grid |
Matrix of panels split by a single categorical vector with different y-axis scale range for each panel
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + facet_grid(Type ~ Treatment, scales = "free_y")
|
Themes
Themes govern the overall style of the graphic.
In particular, they control:
- the look and positioning of the axes (and their ticks, titles and labels)
- the look and positioning of the legends (size,alignment, font, direction)
- the look of plots (spacing and titles)
- the look of panels (background, grid lines)
- the look of panels strips (background, alignment, font)
| Theme | Notes, additional parameters | Example |
_bw |
Black and white theme
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + theme_bw()
|
_classic |
Classic theme
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + theme_classic()
|
_grey |
Grey theme
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + theme_grey()
|
_minimal |
Minimal theme
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + theme_minimal()
|
Along with these pre-fabricated themes, it is possible to create your own theme. This is done via the theme() function.
Each themable element comprises of either a line, rectangle or text. Therefore, they can all be modified via one of the following functions:
- element_blank() - remove the element
- element_line() - set the properties of a line
- element_rect() - set the properties of a rectangle
- element_text() - set the properties of text
library(gridExtra)
ggplot(CO2, aes(y=uptake, x=conc)) + geom_smooth(aes(colour=Type)) + geom_point() +
theme(panel.grid.major = element_blank(), # no major grid lines
panel.grid.minor = element_blank(), # no minor grid lines
panel.background = element_blank(), # no background
panel.border = element_blank(), # no plot border
axis.title.y=element_text(size=15, vjust=0,angle=90), # y-axis title
axis.text.y=element_text(size=12), # y-axis labels
axis.title.x=element_text(size=15, vjust=-2), # x-axis title
axis.text.x=element_text(size=12), # x-axis labels
axis.line = element_line(),
legend.position=c(1,0),
legend.justification=c(1,0),
plot.margin=unit(c(0.5,0.5,2,2),"lines")) # plot margins
Examples
Exploring distributions
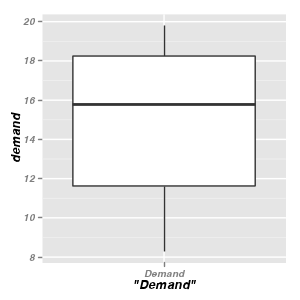
Boxplots - geom_boxplot & stat_boxplot
Univariate boxplots
| Basic boxplot | Plain boxplot |
|
|
|
# Univariate boxplot
ggplot(BOD) + geom_boxplot(aes(y = demand, x = "Demand"))
#Conditional boxplot
p <- ggplot(BOD) +
geom_boxplot(aes(y=demand,x=1)) +
scale_y_continuous("Biochemical oxygen demand (mg/l)") +
scale_x_continuous(limits=c(0,2),breaks=NULL)
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,0.5,2),"lines")
)
|
Conditional (factorial) boxplots
| Basic factorial boxplot | Plain factorial boxplot |
|
|
|
# Conditional boxplot
ggplot(warpbreaks) + geom_boxplot(aes(y = breaks, x = wool))
# Plain conditional boxplot
p <- ggplot(warpbreaks) + geom_boxplot(aes(y = breaks,
x = wool)) + scale_y_continuous("Number of wool breaks") +
scale_x_discrete("Type of wool")
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 12), axis.line = element_line(),
plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"))
|
| Basic factorial boxplot | Plain factorial boxplot |
|
|
|
ggplot(warpbreaks) + geom_boxplot(aes(y = breaks, x = wool,
fill = tension))
p <- ggplot(warpbreaks) +
geom_boxplot(aes(y=breaks,x=wool, fill=tension)) +
scale_y_continuous("Number of wool breaks") +
scale_x_discrete("Type of wool")+
labels=c("Low","Medium","High"),start=0.5,end=1)
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15, vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Violin Plot - geom_violin
| Violin plot | Plain violin plot |
|
|
|
ggplot(warpbreaks, aes(y = breaks, x = wool)) + geom_violin()
library(grid)
library(scales)
p<-ggplot(warpbreaks, aes(y=breaks, x=wool))+
geom_violin()+
scale_x_discrete("Wool type")+
scale_y_continuous("Number of breaks", expand=c(0.05,0), labels=comma)
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,0.2),legend.justification=c(1,0),
plot.margin=unit(c(0.5,0.5,2,2),"lines"),
legend.key=element_blank()
)
|
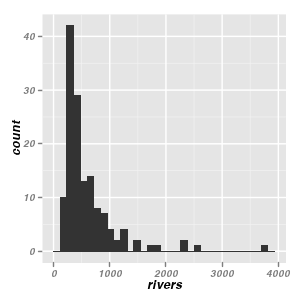
Histograms - geom_histogram, geom_bar & stat_bin
Univariate histograms
| Basic histogram | Plain histogram |
|
|
|
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers))
# OR
ggplot(data = data.frame(rivers)) + geom_histogram(aes(x = rivers))
p <- ggplot(data=data.frame(rivers)) + geom_bar(aes(x=rivers),colour='black',fill='gray')+
scale_x_continuous("Length of rivers (miles)")+
scale_y_continuous("Frequency", expand=c(0,0))+
coord_cartesian(xlim=c(0,4000))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Number of bins | Plain bin width |
|
|
|
# Histogram with customized bin widths
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers),
binwidth = 50)
# OR
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers))
#Plain histogram with custom bin widths
#use the expand() to scale the axis zero to 0
p <- ggplot(data=data.frame(rivers)) + geom_bar(aes(x=rivers),binwidth=50,colour='black',fill='gray')+
scale_x_continuous("Length of rivers (miles)", expand=c(0,0))+
scale_y_continuous("Frequency", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Scaled x-values | Plain transformed x-values |
|
|
|
# Histogram on log transformed data
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers)) +
scale_x_continuous(trans = "log10")
# OR
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers)) +
scale_x_log10()
# Plain histogram of log transformed data
#define a new axis label formattter
p <- ggplot(data=data.frame(rivers)) + geom_bar(aes(x=rivers),colour='black',fill='gray')+
scale_x_continuous("Length of rivers (miles)", expand=c(0,0),trans="log10")+
scale_y_continuous("Frequency", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Scaled x-axis | Plain transformed x- coordinates |
|
|
|
# Histogram of linear data on log transformed axis
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers)) +
coord_trans(xtrans = "log1p")
# Plain histogram of linear data on log transformed axis
#define a new axis label formattter
p <- ggplot(data=data.frame(rivers)) + geom_bar(aes(x=rivers),colour='black',fill='gray')+
scale_x_continuous("Length of rivers (miles)", expand=c(0,0))+
coord_trans(xtrans="log1p")+
scale_y_continuous("Frequency", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Gradient fill | Plain scaled grey gradient fill |
|
|
|
ggplot(data = data.frame(rivers)) + geom_bar(aes(x = rivers,
fill = ..count..))
# Plain histogram with gradient fill
#define a new axis label formattter
p <- ggplot(data=data.frame(rivers)) +
geom_bar(aes(x=rivers,fill=..count..))+
geom_bar(aes(x=rivers, fill=..count..),colour="black",guide=FALSE)+
scale_x_continuous("Length of rivers (miles)", expand=c(0,0))+
scale_y_continuous("Frequency", expand=c(0,0))+
scale_fill_gradient(low="grey90", high="grey40")
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Conditional (factorial) histograms
| Basic histogram | Plain histogram |
|
|
|
ggplot(data = iris) + geom_bar(aes(x = Sepal.Length,
fill = Species), position = "identity")
# OR
ggplot(data = iris) + geom_histogram(aes(x = Sepal.Length,
fill = Species), position = "identity")
#Conditional histogram
p <- ggplot(data=iris) +
geom_bar(aes(x=Sepal.Length, fill=Species), position="identity")+
scale_x_continuous("Sepal length (mm)", expand=c(0,0))+
scale_y_continuous("Frequency", expand=c(0,0))+
scale_fill_grey()
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Basic histogram | Plain histogram |
|
|
|
ggplot(data = iris) + geom_bar(aes(x = Sepal.Length,
fill = Species), position = "dodge")
# OR
ggplot(data = iris) + geom_histogram(aes(x = Sepal.Length,
fill = Species), position = "dodge")
#Transparent Conditional Histogram
p <-ggplot(data=iris)+
geom_bar(aes(x=Sepal.Length,fill=Species), alpha=0.5, stat="bin", position="identity")+
geom_step(aes(x=Sepal.Length, colour=Species,fill=Species),stat="bin", position=position_identity())+#,
scale_x_continuous("Sepal length (mm)", expand=c(0,0))+
scale_y_continuous("Frequency", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
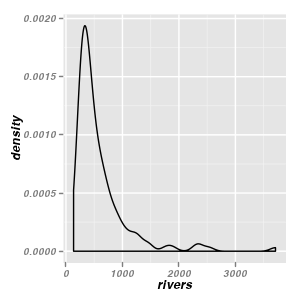
Density plots - geom_density & stat_density
Univariate density
plots
| Basic density plot | Plain density plot |
|
|
|
ggplot(data = data.frame(rivers)) + geom_density(aes(x = rivers))
p <- ggplot(data=data.frame(rivers)) +
geom_density(aes(x=rivers),colour='black',fill='grey90')+
scale_x_continuous("Length of rivers (miles)", expand=c(0,0))+
scale_y_continuous(expression(paste("Density (",phantom() %*% 10^-4,")")), expand=c(0,0),
labels=function(x){format(x*10000,nsmall=1,scientific=FALSE)})
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=12),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Basic smoother density plot | Plain smoother density plot |
|
|
|
ggplot(data = data.frame(rivers)) + geom_density(aes(x = rivers),
adjust = 5)
myF <- function(x) {
format(x * 10000, nsmall = 1, scientific = FALSE)
}
p <- ggplot(data = data.frame(rivers)) + geom_density(aes(x = rivers),
adjust = 5, colour = "black", fill = "grey90") +
scale_x_continuous("Length of rivers (miles)",
expand = c(0, 0)) + scale_y_continuous("Density (/10000)",
expand = c(0, 0), labels = myF)
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 12), axis.line = element_line(),
plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"))
|
| Basic smoother density plot | Plain smoother density plot |
|
|
|
ggplot(data = data.frame(rivers)) + geom_density(aes(x = rivers)) +
scale_x_continuous(trans = "log10")
p <- ggplot(data=data.frame(rivers)) +
geom_density(aes(x=rivers),colour='black',fill='grey90')+
scale_x_continuous("Length of rivers (miles)", expand=c(0,0),
trans="log10", breaks=c(250,500,1000,2000,3000),label=c(250,500,1000,2000,3000))+
scale_y_continuous("Density", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Factorial density
| Basic factorial density plot | Plain factorial density plot |
|
|
|
ggplot(data = iris) + geom_density(aes(x = Sepal.Length,
colour = Species))
# Plain conditional density plot
p <- ggplot(data=iris) + geom_density(aes(x=Sepal.Length, colour=Species))+
scale_x_continuous("Sepal length (mm)", expand=c(0,0))+
scale_y_continuous("Density", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Basic factorial density plot | Plain factorial density plot |
|
|
|
# Conditional density plot
ggplot(data = iris) + geom_density(aes(x = Sepal.Length,
fill = Species))
# Plain conditional density plot
p <- ggplot(data=iris) +
geom_density(aes(x=Sepal.Length,fill=Species), alpha=0.4, colour=NA)+
geom_density(aes(x=Sepal.Length,fill=Species, colour=Species), alpha=0.0, show_guide=FALSE)+
scale_x_continuous("Sepal length (mm)", expand=c(0,0))+
scale_y_continuous("Density", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Line graphs - geom_line
| Basic line graph | Plain line graph |
|
|
|
ggplot(BOD) + geom_line(aes(y = demand, x = Time))
# Plain line plot
p <- ggplot(data=BOD) +
geom_line(aes(y=demand,x=Time),size=2)+
scale_x_continuous("Time (days)", expand=c(0.05,0), limits=c(0,8))+
scale_y_continuous("Demand (mg/l)", expand=c(0.05,0), limits=c(8,20))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Basic line graph | Plain line graph |
|
|
|
ggplot(BOD, aes(y = demand, x = Time)) + geom_line() +
geom_point()
# Plain line plot
p <- ggplot(data=BOD) +
geom_line(aes(y=demand,x=Time),size=2)+
scale_x_continuous("Time (days)", expand=c(0.05,0), limits=c(0,8))+
scale_y_continuous("Demand (mg/l)", expand=c(0.05,0), limits=c(8,20))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Scatterplots -
geom_point, geom_line, geom_smooth, stat_smooth & stat_summary
Simple scatterplots
| Basic scatterplot | Plain scatterplot |
|
|
|
ggplot(BOD) + geom_point(aes(y = demand, x = Time))
# Plain scatterplot
p <- ggplot(data=BOD) +
geom_point(aes(y=demand,x=Time),size=3)+
scale_x_continuous("Time (days)", expand=c(0,0), limits=c(0,8))+
scale_y_continuous("Demand (mg/l)", expand=c(0,0), limits=c(8,20))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Trends and smoothers
| Linear trend | Plain linear trend with
95%
confidence and |
|
|
|
ggplot(BOD) + geom_point(aes(y = demand, x = Time)) +
geom_smooth(aes(y = demand, x = Time), method = "lm")
# fit linear model (in order to get confidence bands)
BOD.lm <- lm(demand~Time, data=BOD)
xs <- seq(min(BOD$Time), max(BOD$Time), l=1000)
BOD.predict <- predict(BOD.lm,
newdata=data.frame(Time=xs),interval='confidence', se=TRUE)
BOD.predict <- data.frame(BOD.predict$fit, se=BOD.predict$se.fit,Time=xs)
# Create a plain scatterplot with smoother and confidence bands
p <- ggplot(data=BOD) +
geom_point(aes(y=demand, x=Time),colour='grey',size=2)+
geom_line(aes(y=demand,x=Time),stat="smooth", method="lm")+
scale_x_continuous("Time (days)", limits=c(1,7))+
scale_y_continuous("Demand (mg/l)")
p <- p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
# add the confidence bands
p+geom_line(data=BOD.predict, aes(y=upr,x=Time), linetype=2)+
geom_line(data=BOD.predict, aes(y=lwr,x=Time), linetype=2)
|
| Loess smoother | Plain loess smoother with standard error |
|
|
|
ggplot(BOD) + geom_point(aes(y = demand, x = Time)) +
geom_smooth(aes(y = demand, x = Time), method = "loess",
degree = 1, se = TRUE)
# Fit a loess smoother
BOD.loess <- loess(demand~Time, data=BOD, degree=1)
xs <- seq(min(BOD$Time), max(BOD$Time), l=1000)
BOD.predict <- predict(BOD.loess,newdata=data.frame(Time=xs), se=TRUE)
BOD.predict <- with(BOD.predict,data.frame(fit,lwr=fit-se.fit,upr=fit+se.fit,Time=xs))
# Plain scatterplot with loess smoother and confidence bands
p <- ggplot(data=BOD) +
geom_point(aes(y=demand, x=Time),colour='grey',size=2)+
geom_line(data=BOD.predict,aes(y=fit,x=Time))+
scale_x_continuous("Time (days)", limits=c(1,7))+
scale_y_continuous("Demand (mg/l)")
p <- p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
# add confidence bands
p+geom_line(data=BOD.predict, aes(y=upr,x=Time), linetype=2)+
geom_line(data=BOD.predict, aes(y=lwr,x=Time), linetype=2)
|
| Generalized additive model (GAM) | Plain gam with 95% CI
based on
1.96 (dashed; ggplot default) and qt(0.975,df) |
|
|
|
library(mgcv)
ggplot(CO2, aes(y = uptake, x = conc)) + geom_point() +
stat_smooth(geom = "smooth", method = "gam", formula = y ~
s(x, k = 6))
#manually fit a GAM
library(mgcv)
CO2.gam <- gam(uptake~s(conc,k=6), data=CO2)
xs <- seq(min(CO2$conc), max(CO2$conc), l=1000)
CO2.predict <- predict(CO2.gam,newdata=data.frame(conc=xs),se.fit=TRUE)
df <- sum(CO2.gam$edf[-1])
#generate 95% CI predictions based on 1.96SE and degrees of freedom
CO2.predict1 <-with(CO2.predict,data.frame(fit,lwr=fit-(1.96*se.fit),
upr=fit+(1.96*se.fit),conc=xs))
CO2.predict2 <- with(CO2.predict,data.frame(fit,lwr=fit-(qt(0.975,df)*se.fit),
upr=fit+(qt(0.975,df)*se.fit),conc=xs))
p <- ggplot(data=CO2) +
geom_point(aes(y=uptake, x=conc),colour='grey',size=2)+
#stat_smooth(aes(y=uptake, x=conc),geom="smooth",method="gam",formula=y~s(x,k=6))+
scale_x_continuous(expression(paste("Ambient ",CO[2]," concentration (mg/l)", sep="")))+
scale_y_continuous(expression(paste(CO[2]," uptake rate (",mu*mol/m^2/sec,")", sep="")))
p <- p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
# add the trendline and confidence bands
p+geom_line(data=CO2.predict1, aes(y=fit,x=conc),)+
geom_line(data=CO2.predict1, aes(y=upr,x=conc), linetype=2)+
geom_line(data=CO2.predict1, aes(y=lwr,x=conc), linetype=2)+
geom_line(data=CO2.predict2, aes(y=upr,x=conc), linetype=3)+
geom_line(data=CO2.predict2, aes(y=lwr,x=conc), linetype=3)
|
| Means plot | Plain gam with 95% CI
based on
1.96 (dashed; ggplot default) and qt(0.975,df) |
|
|
|
p <- ggplot(CO2, aes(y = uptake, x = conc)) + geom_pointrange(stat = "summary",
fun.data = "mean_cl_boot")
p
p <- ggplot(data=CO2, aes(y=uptake, x=conc)) +
scale_x_continuous(expression(paste("Ambient ",CO[2]," concentration (mg/l)", sep="")))+
scale_y_continuous(expression(paste(CO[2]," uptake rate (",mu*mol/m^2/sec,")", sep="")))
p <- p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
# add the errorbars
p+geom_pointrange(stat="summary", fun.data="mean_cl_normal")
|
| Means plot | Plain gam with 95% CI
based on
1.96 (dashed; ggplot default) and qt(0.975,df) |
|
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_errorbar(stat = "summary",
fun.data = "mean_cl_boot") + geom_point(stat = "summary",
fun.y = "mean")
p <- ggplot(data=CO2, aes(y=uptake, x=conc)) +
stat_smooth(geom="smooth",method="gam",formula=y~s(x,k=6),se=FALSE, colour="gray")+
geom_errorbar(stat="summary", fun.data="mean_cl_boot") +geom_point(stat="summary",fun.y="mean")+
scale_x_continuous(expression(paste("Ambient ",CO[2]," concentration (mg/l)", sep="")))+
scale_y_continuous(expression(paste(CO[2]," uptake rate (",mu*mol/m^2/sec,")", sep="")))
p +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Bargraphs (dynamite plots) - geom_bar & stat_summary
Simple bargraphs
| Bargraph | Plain bargraph |
|
|
|
ggplot(warpbreaks, aes(y = breaks, x = tension)) +
geom_bar(stat = "summary", fun.y = mean) + geom_errorbar(stat = "summary",
fun.data = "mean_cl_normal", width = 0.1)
p <- ggplot(data=warpbreaks, aes(y=breaks, x=tension)) +
geom_bar(stat="summary", fun.y=mean,color="black",fill="grey80")+
geom_errorbar(stat="summary", fun.data="mean_cl_normal", width=0.1)+
scale_x_discrete("Tension")+
scale_y_continuous("Number of breaks", expand=c(0,0))
p + theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Conditional (factorial) bargraphs
| Conditional bargraph | Plain conditional bargraph |
|
|
|
ggplot(warpbreaks, aes(y = breaks, x = tension, group = wool)) +
geom_bar(aes(fill = wool), position = position_dodge(0.9),
stat = "summary", fun.y = mean) + geom_errorbar(position = position_dodge(0.9),
stat = "summary", fun.data = "mean_cl_normal",
width = 0.1)
p <- ggplot(data = warpbreaks, aes(y = breaks, x = tension,
group = wool)) + geom_bar(aes(fill = wool), position = "dodge",
stat = "summary", fun.y = mean) + geom_bar(aes(fill = wool),
position = "dodge", stat = "summary", fun.y = mean,
color = "black", show_guide = FALSE) + scale_fill_grey("Wool type") +
geom_errorbar(position = position_dodge(0.9), stat = "summary",
fun.data = "mean_cl_normal", width = 0.1) +
scale_x_discrete("Tension") + scale_y_continuous("Number of wool breaks",
expand = c(0, 0))
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 10), axis.line = element_line(),
legend.position = c(1, 1), legend.justification = c(1,
1), plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"))
|
Bar charts - geom_bar
| Stacked barchart | Plain stacked barchart |
|
|
|
# based on pre-calculated counts
warpbreaks.c <- ddply(warpbreaks, ~wool + tension,
function(x) data.frame(count = sum(x$breaks)))
ggplot(warpbreaks.c, aes(x = tension, y = count, fill = wool)) +
geom_bar(stat = "identity") + ylab("Number of breaks")
#based on pre-calculated counts
warpbreaks.c<-ddply(warpbreaks,~wool+tension, function(x) data.frame(count=sum(x$breaks)))
p <- ggplot(warpbreaks.c, aes(x=tension,y=count,fill=wool))+
geom_bar(aes(fill=wool), stat='identity')+
geom_bar(aes(fill=wool), stat='identity', colour="black",show_guide=FALSE)+
scale_fill_grey("Wool type")+
scale_x_discrete("Tension")+
scale_y_continuous("Number of wool breaks", expand=c(0,0))
p +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
| Horizontal stacked barchart | Plain horizontal stacked barchart |
|
|
|
# based on pre-calculated counts
warpbreaks.c <- ddply(warpbreaks, ~wool + tension,
function(x) data.frame(count = sum(x$breaks)))
ggplot(warpbreaks.c, aes(x = tension, y = count, fill = wool)) +
geom_bar(stat = "identity") + ylab("Number of breaks") +
coord_flip()
#based on pre-calculated counts
warpbreaks.c<-ddply(warpbreaks,~wool+tension, function(x) data.frame(count=sum(x$breaks)))
p <- ggplot(warpbreaks.c, aes(x=tension,y=count,fill=wool))+ coord_flip()+
geom_bar(aes(fill=wool), stat='identity')+
geom_bar(aes(fill=wool), stat='identity', colour="black",show_guide=FALSE)+
scale_fill_grey("Wool type")+
scale_x_discrete("Tension")+
scale_y_continuous("Number of wool breaks", expand=c(0.05,0))
p +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_blank(),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
axis.line = element_line(),
legend.position=c(1,1),legend.justification=c(1,1),
plot.margin=unit(c(0.5,0.5,2,2),"lines")
)
|
Interaction plots - geom_point, geom_line, geom_smooth
| Interaction plot | Plain interaction plot |
|
|
|
ggplot(ToothGrowth, aes(y = len, x = dose, colour = supp)) +
geom_point() + geom_smooth(method = "lm")
p <- ggplot(ToothGrowth, aes(y = len, x = dose, linetype = supp)) +
geom_point(aes(shape = supp)) + geom_smooth(method = "lm") +
scale_linetype_manual(name = "Suppliment type",
values = c(1, 2), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_shape_manual(name = "Suppliment type",
values = c(21, 16), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_x_continuous("Dose (mg)",
labels = comma) + scale_y_continuous("Tooth length (mm)",
expand = c(0.05, 0), labels = comma)
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 10), axis.line = element_line(),
legend.position = c(1, 0), legend.justification = c(1,
0), plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"))
|
| Interaction plot with errorbars | Plain interaction plot with errorbars |
|
|
|
ggplot(ToothGrowth, aes(y = len, x = dose, colour = supp)) +
geom_errorbar(stat = "summary", fun.data = "mean_cl_boot") +
geom_point(stat = "summary", fun.y = "mean") +
geom_line(stat = "summary", fun.y = "mean")
p <- ggplot(ToothGrowth, aes(y = len, x = dose, group = supp)) +
geom_errorbar(stat = "summary", fun.data = "mean_cl_boot",
width = 0.05) + geom_line(aes(linetype = supp),
stat = "summary", fun.y = "mean") + geom_point(aes(shape = supp,
fill = supp), stat = "summary", fun.y = "mean") +
scale_shape_manual(name = "Suppliment type", values = c(21,
16), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_fill_manual(name = "Suppliment type",
values = c("white", "black"), breaks = c("OJ",
"VC"), labels = c("Orange juice", "Vitamine C")) +
scale_linetype_manual(name = "Suppliment type",
values = c(1, 2), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_x_continuous("Dose (mg)",
labels = comma) + scale_y_continuous("Tooth length (mm)",
expand = c(0.05, 0), labels = comma)
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 10), axis.line = element_line(),
legend.position = c(1, 0), legend.justification = c(1,
0), plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"),
legend.key = element_blank())
|
| Interaction plot with dodge | Plain interaction plot with dodge |
|
|
|
ggplot(ToothGrowth, aes(y = len, x = dose, colour = supp)) +
geom_errorbar(stat = "summary", fun.data = "mean_cl_boot",
position = position_dodge(0.2)) + geom_point(stat = "summary",
fun.y = "mean", position = position_dodge(0.2)) +
geom_line(stat = "summary", fun.y = "mean", position = position_dodge(0.2))
p <- ggplot(ToothGrowth, aes(y = len, x = dose, group = supp)) +
geom_errorbar(stat = "summary", fun.data = "mean_cl_boot",
width = 0.05, position = position_dodge(0.2)) +
geom_line(aes(linetype = supp), stat = "summary",
fun.y = "mean", position = position_dodge(0.2)) +
geom_point(aes(shape = supp, fill = supp), size = 3,
stat = "summary", fun.y = "mean", position = position_dodge(0.2)) +
scale_shape_manual(name = "Suppliment type", values = c(21,
16), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_fill_manual(name = "Suppliment type",
values = c("white", "black"), breaks = c("OJ",
"VC"), labels = c("Orange juice", "Vitamine C")) +
scale_linetype_manual(name = "Suppliment type",
values = c(1, 2), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_x_continuous("Dose (mg)",
labels = comma) + scale_y_continuous("Tooth length (mm)",
expand = c(0.05, 0), labels = comma)
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 10), axis.line = element_line(),
legend.position = c(1, 0), legend.justification = c(1,
0), plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"),
legend.key = element_blank())
|
Scatterplot matrix - plotmatrix
| Means plot | Plain gam with 95% CI
based on
1.96 (dashed; ggplot default) and qt(0.975,df) |
|
|
|
p <- plotmatrix(iris) + geom_point(aes(colour = Species))
|
|
library(grid)
library(scales)
p <- ggplot(ToothGrowth, aes(y = len, x = dose, group = supp)) +
geom_errorbar(stat = "summary", fun.data = "mean_cl_boot",
width = 0.05, position = position_dodge(0.2)) +
geom_line(aes(linetype = supp), stat = "summary",
fun.y = "mean", position = position_dodge(0.2)) +
geom_point(aes(shape = supp, fill = supp), size = 3,
stat = "summary", fun.y = "mean", position = position_dodge(0.2)) +
scale_shape_manual(name = "Suppliment type", values = c(21,
16), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + scale_fill_manual(name = "Suppliment type",
values = c("white", "black"), breaks = c("OJ",
"VC"), labels = c("Orange juice", "Vitamine C")) +
scale_linetype_manual(name = "Suppliment type",
values = c(1, 2), breaks = c("OJ", "VC"), labels = c("Orange juice",
"Vitamine C")) + # scale_fill_grey('Wool type')+
scale_x_continuous("Dose (mg)", label = comma) + scale_y_continuous("Tooth length (mm)",
expand = c(0.05, 0), label = comma)
p + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
panel.background = element_blank(), panel.border = element_blank(),
axis.title.y = element_text(size = 15, vjust = 0,
angle = 90), axis.text.y = element_text(size = 12),
axis.title.x = element_text(size = 15, vjust = -1),
axis.text.x = element_text(size = 10), axis.line = element_line(),
legend.position = c(1, 0.1), legend.justification = c(1,
0), plot.margin = unit(c(0.5, 0.5, 2, 2), "lines"),
legend.key = element_blank())
|
Multi-panel (facetted) plot - facet_grid & facet_wrap
2D grid of panels - facet_grid
| Grid of panels | Plain grid of panels |
|
|
|
ggplot(CO2, aes(y = uptake, x = conc)) + geom_smooth() +
geom_point() + facet_grid(Type ~ Treatment)
|
library(grid)
library(scales)
# Create a new instance of the dataset to facilitate more informative panel titles
CO2.a <- CO2
# Re-define the factor labels
CO2.a$Treatment <- factor(CO2.a$Treatment, labels=c(expression(paste("Non-Chilled ", (symbol("\076")*15 * degree * C), sep="")),
expression(paste("Chilled ",(symbol("\074")*5 * degree * C), sep=""))))
CO2.a$Type <- factor(CO2.a$Type, labels=c("Origin:Quebec", "Origin:Mississippi"))
p<-ggplot(CO2.a,aes(y=uptake,x=conc))+
geom_ribbon(aes(ymin=..ymin.., ymax=..ymax..),linetype=2,fill="transparent",colour="black",stat='smooth',method='loess')+
geom_smooth(se=FALSE)+
geom_point()+
facet_grid(Type~Treatment, labeller=label_parsed)+
scale_x_continuous(expression(paste("Ambient ",CO[2]," concentration (mg/l)", sep="")))+
scale_y_continuous(expression(paste(CO[2]," uptake rate (",mu*mol/m^2/sec,")", sep="")))
p +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
panel.border = element_rect(fill="transparent",colour="black"),
axis.title.y=element_text(size=15, vjust=0,angle=90),
axis.text.y=element_text(size=12),
axis.title.x=element_text(size=15,vjust=-1),
axis.text.x=element_text(size=10),
#axis.line = element_line(),
strip.background=element_rect(fill="transparent", colour="black"),
#legend.position=c(1,0.2),legend.justification=c(1,0),
plot.margin=unit(c(0.5,0.5,2,2),"lines"),
legend.key=element_blank()
)
|
Multiple graphs per graphic
Viewports
| Grid of panels | Plain grid of panels |
|
|
|
p1 <- ggplot(CO2, aes(y = uptake, x = Treatment, fill = Type)) + geom_boxplot()
p2 <- ggplot(CO2, aes(y = uptake, x = conc, colour = Type)) + geom_smooth()
p3 <- ggplot(CO2, aes(x = uptake, fill = Type)) + geom_density(alpha = 0.4) + facet_grid(~Treatment)
grid.newpage()
pushViewport(viewport(layout = grid.layout(4, 5)))
pushViewport(viewport(layout.pos.col = 1:2, layout.pos.row = 1:2))
print(p1 + theme(legend.position = "none"), newpage = FALSE)
popViewport(1)
pushViewport(viewport(layout.pos.col = 3:4, layout.pos.row = 1:2))
print(p2 + theme(legend.position = "none"), newpage = FALSE)
popViewport(1)
pushViewport(viewport(layout.pos.col = 1:4, layout.pos.row = 3:4))
print(p3 + theme(legend.position = "none"), newpage = FALSE)
popViewport(1)
library(gridExtra)
tmp <- ggplot_gtable(ggplot_build(p3))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
# legend <- gTree(legend, vp=viewport(layout.pos.col=5, layout.pos.row=1:4)) using grid.arrange for
# convenience could also manually push viewports grid.arrange(arrangeGrob(p3 +
# theme(legend.position='none'), p2 + theme(legend.position='none'), main ='this is a title', left =
# 'This is my global Y-axis title'), legend, widths=unit.c(unit(1, 'npc') - legend$width,
# legend$width), nrow=1)
pushViewport(viewport(layout.pos.col = 5, layout.pos.row = 1:4))
# print(p3+theme(keep='legend_box'), newpage=FALSE) print(p3+theme(keep='legend_box'), newpage=FALSE)
grid.draw(legend)
popViewport(0)
|
#Create a new plain element
element_plain <- function (base_size = 12,base_family=""){ #
structure(list(
axis.line = element_line(),
#axis.text.x = element_text(family = base_family, size = base_size * 0.8, vjust = 1, lineheight = 0.9),
#axis.text.y = element_text(family = base_family, size = base_size * 0.8, hjust = 1, lineheight = 0.9),
#axis.ticks = element_line(colour = "black", size = 0.2),
#axis.title.x = element_text(family = base_family, size = base_size, vjust = 0.5),
#axis.title.y = element_text(family = base_family, size = base_size, vjust = 0.5, angle = 90),
#axis.ticks.length = unit(0.15, "cm"),
#axis.ticks.margin = unit(0.1, "cm"),
#legend.background=element_blank(),
#legend.margin=unit(0.2,"cm"),
#legend.key = element_rect(colour = "grey80"),
#legend.key.size=unit(1.2,"lines"),
#legend.key.height=NULL,
#legend.key.width=NULL,
#legend.text=element_text(family = base_family, size = base_size * 0.8),
#legend.text.align=NULL,
#legend.title=element_text(family = base_family, face = "bold", size = base_size * 0.8, hjust = 0),
#legend.title.align=NULL,
#legend.position = "right",
#legend.direction=NULL,
#legend.justification="center",
#legend.box=NULL,
#panel.background = element_blank(),
#panel.border = element_blank(),
#panel.grid.major = element_blank(),
#panel.grid.minor = element_blank(),
#panel.margin = unit(0.25, "lines"),
#strip.background=element_rect(fill="transparent", colour="black"),
#strip.text.x=element_text(family = base_family, size = base_size * 0.8),
#strip.text.y=element_text(family = base_family, size = base_size * 0.8, angle = -90),
#plot.background = element_blank(),
plot.margin=unit(c(0.5,0.5,1,1),"lines"),
plot.title=element_blank()
), class = "theme")
}
#Construct the boxplots
p1 <- ggplot(CO2, aes(y=uptake,x=Treatment,fill=Type))+
geom_boxplot(alpha=0.4)+scale_fill_manual(values=c("white","grey"))+
element_plain()
p2 <- ggplot(CO2, aes(y=uptake, x=conc, linetype=Type))+
geom_smooth(color="black",se=FALSE)+
geom_smooth(color="black", show_guide=FALSE)+
scale_linetype()+
element_plain()
p3<-ggplot(CO2, aes(x=uptake, fill=Type)) +
geom_density(alpha=0.4, colour=NA)+
geom_density(alpha=0, show_guide=FALSE)+
facet_grid(~Treatment)+
scale_fill_manual(values=c("white","grey"))+
element_plain()
library(gridExtra)
tmp <- ggplot_gtable(ggplot_build(p3))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
tmp <- ggplot_gtable(ggplot_build(p2))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend1 <- tmp$grobs[[leg]]
##legend <- gTree(legend, vp=viewport(layout.pos.col=5, layout.pos.row=1:4))
grid.newpage()
pushViewport(viewport(layout=grid.layout(4,5)))
pushViewport(viewport(layout.pos.col=1:2, layout.pos.row=1:2))
print(p1+theme(legend.position="none"), newpage=FALSE)
popViewport(1)
pushViewport(viewport(layout.pos.col=3:4, layout.pos.row=1:2))
print(p2+theme(legend.position="none"), newpage=FALSE)
popViewport(1)
pushViewport(viewport(layout.pos.col=1:4, layout.pos.row=3:4))
print(p3+theme(legend.position="none"), newpage=FALSE)
popViewport(1)
pushViewport(viewport(layout.pos.col=5, layout.pos.row=1:4))
grid.draw(legend)
grid.draw(legend1)
popViewport(0)
|
- axis ticks etc
Welcome to the end of this tutorial




























































































































































































