Tutorial 6.2a - t-tests
21 May 2017
Two independent sample t-tests
As a starting point for this two sample t-tests, we will generate some fabricated representing a single response collected from two populations (Population A and Population B). The characteristics of the populations (which are obviously not normally known - rather these are the parameters that we seek to estimate and make inferences about) as well as some aspects of the experimental design used to collect the observations are:
- the mean of Population 1 is 105
- the mean of Population 2 is 77.5
- both populations have a standard deviation of 3.0
- Population 1 and 2 were sampled with $n=60$ and $n=40$ respectively
set.seed(1) nA <- 60 #sample size from Population A nB <- 40 #sample size from Population B muA <- 105 #population mean of Population A muB <- 77.5 #population mean of Population B sigma <- 3.0 #standard deviation of both populations (equally varied) yA <- rnorm(nA, muA, sigma) #Population A sample yB <- rnorm(nB, muB, sigma) #Population B sample y <- c(yA, yB) x <- factor(rep(c("A", "B"), c(nA, nB))) #categorical listing of the populations data <- data.frame(y, x) # dataset
Assumptions and exploratory data analysis
We are going to use the samples to estimate the population means as well as test the null hypothesis that the populations are equal (i.e. that the population means are the same - Population A minus Population b equals 0).
This simple null hypothesis can be tested using a t-test. However, for the test to be reliable, it assumes that:
- the populations from which the samples were collected were normally distributed
- the populations from which the samples were collected were equally varied
- the collected samples represented the populations in an unbiased manner
The last of these assumptions can only be addressed at the design and data collection phase. These fabricated data were generated from equally varied normal distributions and thus should adhere to the other two assumptions. Nevertheless, boxplots are a useful diagnotic.
boxplot(y~x, data)
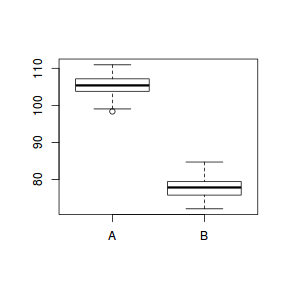
tapply(data$y, data$x,var)
A B 6.581824 8.474237
In R, pooled-variances (student) and separate variances (Welch test) t-test are performed using the t.test() function.
- For pooled-variances t-test:
t.test(y~x, data, var.equal=TRUE)
Two Sample t-test data: y by x t = 49.727, df = 98, p-value < 2.2e-16 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: 26.39339 28.58754 sample estimates: mean in group A mean in group B 105.32285 77.83238 - For separate-variances t-test:
t.test(y~x, data, var.equal=FALSE)
Welch Two Sample t-test data: y by x t = 48.479, df = 76.318, p-value < 2.2e-16 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: 26.36115 28.61978 sample estimates: mean in group A mean in group B 105.32285 77.83238
The 95% confidence intervals indicate 95% of such intervals (intervals spanning a sample difference of 26.36-28.59 units) would contain the true mean.
Paired samples t-test
Lets generate some more fabricated data of a single response collected from pairs of samples representing sites sampled before and after an impact. In this case, there is actually only a single population (the difference between before and after). The degree to which sites respond to the impact differs from site to site (the variability) The characteristics of the populations and samples are:
- the mean before response value is 100
- the mean effect on this response after the impact is +30
- the variability of the effect is 5.0
- the variability of the sites is 20
- There were $n=25$ sites before and after the impact
set.seed(1) n.sites <- 25 n.repeats <- 2 n <- n.sites * n.repeats before <- 100 site.sd <- 20 ba.effect <- 30 sigma <- 15 int.effects <- rnorm(n=n.sites, mean=before, sd=site.sd) ba <- gl(2,1,n, lab=c("Before","After")) ba.effects <- rep(ba.effect,n.sites) sites <- gl(n = n.sites, k = n.repeats) all.effects <- c(int.effects, ba.effects) Xmat <- model.matrix(~-1+sites*ba-ba) lin.pred <- Xmat[,] %*% all.effects eps <- rnorm(n=n,mean=0, sd=sigma) y <- lin.pred + eps data1 <- data.frame(y,sites,ba) #check some of the properties #before and after means with(data1, tapply(y,ba,mean))
Before After 104.2735 135.4377
#site standard deviation sqrt(var(with(data1, tapply(y,sites,mean))))
[1] 21.32765
#traditional wide format library(reshape) data2 <- cast(data1,sites~ba, value="y")
Assumptions and exploratory data analysis
A paired t-test is essentially a one sample t-test of the differences between each pair (the null hypothesis is that the mean difference equals zero). Therefore, the regular assumptions pertain to the differences
Using the repeated measures (wide) data format, we create a new column that is the difference between the before and after columns - this mimics the response.
data2<-within(data2, Diff <- After-Before)
As with a regular t-test, normality can be explored using boxplots.
boxplot(data2$Diff)
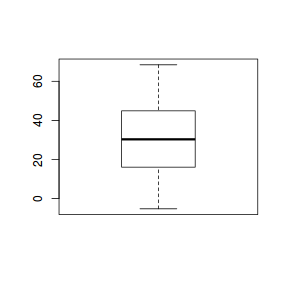
In R, paired t-test are also performed using the t.test() function.
t.test(data2$After, data2$Before, paired=TRUE)
Paired t-test
data: data2$After and data2$Before
t = 7.6622, df = 24, p-value = 6.714e-08
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
22.76984 39.55870
sample estimates:
mean of the differences
31.16427
Worked examples
- Logan (2010) - Chpt 6
- Quinn & Keough (2002) - Chpt 6
Hypothesis testing
Furness & Bryant (1996) studied the energy budgets of breeding northern fulmars (Fulmarus glacialis) in Shetland. As part of their study, they recorded the body mass and metabolic rate of eight male and six female fulmars.
Download Furness data set| Format of furness.csv data files | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 |
||||||||||||||||||||||||
Open the furness data file.
furness <- read.csv('../downloads/data/furness.csv',header=T, strip.white=TRUE) head(furness)
SEX METRATE BODYMASS 1 Male 2950.0 875 2 Female 1956.1 635 3 Male 2308.7 765 4 Male 2135.6 780 5 Male 1945.6 790 6 Female 1490.5 635
-
The researchers were interested in testing whether there is a difference in the metabolic rate of male and female breeding northern fulmars. In light of this, list the following:
- The biological hypotheses of interest
- The biological null hypotheses
- The statistical null hypotheses (H0)
- The biological hypotheses of interest
- For the null hypothesis test of interest (that the mean population metabolic rate of males and females were the same), calculate the
Degrees of freedom
Show code#number of replicates in each group minus 2 (one for each population) sum(with(furness, tapply(METRATE, SEX, length)))-2
[1] 12
-
Calculate the critical t-values for the following null hypotheses (&alpha = 0.05)
- The metabolic rate of males is higher than that females
(one-tailed test)
HINT
- The metabolic rate of males is the same as that of females
(two-tailed test)
HINT
Show codeqt(0.05,df=12,lower.tail=F)
[1] 1.782288
qt(0.05/2,df=12,lower.tail=F)
[1] 2.178813
- The metabolic rate of males is higher than that females
(one-tailed test)
HINT
-
In the table below, list the assumptions of a
t-test along with how violations of each assumption are diagnosed and/or the
risks of violations are minimized.
Assumption Diagnostic/Risk Minimization I. II. III. So, we wish to investigate whether or not male and female fulmars have the same metabolic rates, and that we intend to use a t-test to test the null hypothesis that the population mean metabolic rate of males is equal to the population mean metabolic rate of females. Having identified the important assumptions of a t-test, use the samples to evaluate whether the assumptions are likely to be violated and thus whether a t-test is likely to be reliability.
-
Is there any evidence that; HINT
- The assumption of normality has been violated?
- The assumption of homogeneity of variance has been violated?
Show codeboxplot(METRATE~SEX, data=furness)

- The assumption of normality has been violated?
-
Perform a t-test to examine the effect of sex on the mass of fulmars using either (which ever is most appropriate) a
pooled variance t-test
(for when population variances are very similar HINT) or
separate variance t-test
(for when the variance of one population is likely to be up to 2.5 times greater or less than the other population HINT). Ensure that you are familiar with the
output of a t-test.
- What is the t-value? (Excluding the sign. The sign will depend on whether you compared males to females or females to males, and thus only indicates which group had the higher mean).
- What is the df (degrees of freedom).
- What is the p value.
Show code#Separate variance t-test is probably more reliable, as there is some evidence that the two populations are not exactly equal in variability t.test(METRATE~SEX, equal.var=F,data=furness)
Welch Two Sample t-test data: METRATE by SEX t = -0.77317, df = 10.468, p-value = 0.4565 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -1075.3208 518.8042 sample estimates: mean in group Female mean in group Male 1285.517 1563.775 - What is the t-value? (Excluding the sign. The sign will depend on whether you compared males to females or females to males, and thus only indicates which group had the higher mean).
-
Write the results out as though you were writing a research paper/thesis. For example (select the phrase that applies and fill in gaps with your results):
The mean metabolic rate of male fulmars was (choose correct option)
(t =
, df =
, P =
) the mean metabolic rate of female fulmars.
- Construct a
bar graph
showing the mean metabolic rate of male and female fulmars and an indication of the precision of the means with error bars.
Show code
# calculate the means means <- tapply(furness$METRATE, furness$SEX, mean) # calculate the standard deviation sds <- tapply(furness$METRATE, furness$SEX, sd) # calculate the lengths ns <- tapply(furness$METRATE, furness$SEX, length) # calculate the standard errors ses <- sds/sqrt(ns) # plot the bars xs <- barplot(means,beside=T) # load package containing error bar function library(Hmisc) # plot the error bars errbar(xs, means, means+ses, means-ses, add=T)
 Show code
Show code# set the size of the figure margins opar<-par(mar=c(4,5,0,0)) # calculate the means means <- tapply(furness$METRATE, furness$SEX, mean) # calculate the standard deviation sds <- tapply(furness$METRATE, furness$SEX, sd) # calculate the lengths ns <- tapply(furness$METRATE, furness$SEX, length) # calculate the standard errors ses <- sds/sqrt(ns) #calculate the data limits lim <- max(means+ses) # plot the bars xs <- barplot(means,beside=T, axes=FALSE, ann=FALSE, ylim=c(0,lim)) # plot the error bars arrows(xs,means-ses,xs,means+ses, len=0.1, ang=90, code=3) # generate the axes mtext("Sex",1, line=2.5, cex=2) axis(2,las=1) mtext("Metabolic rate",2,line=3.5, cex=2) box(bty="l")
 Show ggplot code
Show ggplot codelibrary(ggplot2) library(plyr) library(gmodels) dat <- ddply(furness, ~SEX, function(x) { data.frame(Metrate=mean(x$METRATE), t(ci(x$METRATE))) }) head(dat)
SEX Metrate Estimate CI.lower CI.upper Std..Error 1 Female 1285.517 1285.517 843.7436 1727.290 171.8572 2 Male 1563.775 1563.775 816.0607 2311.489 316.2085
ggplot(dat, aes(y=Metrate, x=SEX)) + geom_point(data=furness, aes(y=METRATE, x=SEX), color='grey')+ geom_pointrange(aes(ymin=CI.lower, ymax=CI.upper))+ scale_y_continuous(expression(Metabolic~rate~(HJ/day)))+ scale_x_discrete('')+ theme_classic()+theme(axis.title.y=element_text(vjust=2, size=rel(1.25), face='bold'))

The appropriate statistical test for testing the null hypothesis that the means of two independent populations are equal is a t-test
Before proceeding, make sure you understand what is meant by normality and equal variance as well as the principles of hypothesis testing using a t-test.
Since most hypothesis tests follow the same basic procedure, confirm that you understand the basic steps of hypothesis tests
Paired data
Here is a modified example from Quinn and Keough (2002). Elgar et al. (1996) studied the effect of lighting on the web structure or an orb-spinning spider. They set up wooden frames with two different light regimes (controlled by black or white mosquito netting), light and dim. A total of 17 orb spiders were allowed to spin their webs in both a light frame and a dim frame, with six days `rest' between trials for each spider, and the vertical and horizontal diameter of each web was measured. Whether each spider was allocated to a light or dim frame first was randomized. The H0's were that each of the two variables (vertical diameter and horizontal diameter of the orb web) were the same in dim and light conditions. Elgar et al. (1996) correctly treated these as paired comparisons because the same spider spun her web in a light frame and a dark frame.
Download Elgar data set| Format of elgar.csv data files | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
 | ||||||||||||||||||||||||||||||
Open the elgar data file.
elgar <- read.csv("../downloads/data/elgar.csv", header=T, strip.white=T) head(elgar)
PAIR VERTDIM HORIZDIM VERTLIGHT HORIZLIGHT 1 K 300 295 80 60 2 M 240 260 120 140 3 N 250 280 170 160 4 O 220 250 90 120 5 P 160 160 150 180 6 R 170 150 110 90
- What is an appropriate statistical test for testing an hypothesis about the difference in dimensions of webs spun in light versus dark conditions? Explain why?
- The actual H0 is that the mean of the differences between the pairs (light and dim for each spider) equals zero. Use a
paired t-test
to test the H0 that the mean of the differences in vertical diameter (HINT) and separately, in horizontal diameter (HINT) of the web between the pairs (light and dim for each spider) equal zero.
Show code
t.test(elgar$VERTDIM,elgar$VERTLIGHT, paired=T)
Paired t-test data: elgar$VERTDIM and elgar$VERTLIGHT t = 0.96545, df = 16, p-value = 0.3487 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -24.61885 65.79532 sample estimates: mean of the differences 20.58824t.test(elgar$HORIZDIM,elgar$HORIZLIGHT, paired=T)
Paired t-test data: elgar$HORIZDIM and elgar$HORIZLIGHT t = 2.1482, df = 16, p-value = 0.04735 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: 0.6085725 91.7443687 sample estimates: mean of the differences 46.17647 -
Write the results out as though you were writing a research paper/thesis. For example (select the phrase that applies and fill in gaps with your results):
The mean vertical diameter of spider webs in dim conditions was (choose correct option)
(t =
, df =
, P =
)
the vertical dimensions in light conditions.
The mean horizontal diameter of spider webs in dim conditions was (choose correct option)
(t =
, df =
, P =
)
the horizontal dimensions in light conditions.
Non-parametric tests
We will now revisit the data set of Furness & Bryant (1996) that was used in Question 4 to investigate the effects of gender on the metabolic rates of breeding northern fulmars (Fulmarus glacialis). Furness & Bryant (1996) also recorded the body mass of the eight male and six female fulmars they captured.
Since the males and female fulmars were all independent of one another, a t-test would be appropriate to test the null hypothesis of no difference in mean body weight of male and female fulmars.
-
Are the assumptions underlying this test met? (Y or N) Hint: check the relative sizes of the two sample variances and the distribution of body weight for each sex. Show code
boxplot(BODYMASS~SEX, data=furness)

tapply(furness$BODYMASS,furness$SEX,mean)
Female Male 643.00 839.75
tapply(furness$BODYMASS,furness$SEX,var)
Female Male 166.000 6214.786
-
The Wilcoxon-Mann-Whitney test is described as a non-parametric test for comparing two groups.
- What null hypothesis does this test actually evaluate?
- What are the underlying assumptions of a Wilcoxon-Mann-Whitney test?
- What null hypothesis does this test actually evaluate?
-
If the assumptions are met, test the null hypothesis of no difference in body weight between male
and female fulmars using a
Wilcoxon test
HINT. Based on this outcome, what are your conclusions?
- Statistical:
- Biological
(include trend):
Show codewilcox.test(BODYMASS~SEX, data=furness)
Wilcoxon rank sum test with continuity correction data: BODYMASS by SEX W = 0, p-value = 0.002309 alternative hypothesis: true location shift is not equal to 0
- Statistical:
- Construct a
bar graph
showing the mean mass of male and female fulmars and an indication of the precision of the means with error bars.
Show code
##use ggplot to calculate bootstrap confidence intervals and means ggplot(furness, aes(y=METRATE, x=SEX)) + geom_point(data=furness, aes(y=METRATE, x=SEX), color='grey')+ geom_pointrange(stat='summary', fun.data='mean_cl_boot')+ scale_y_continuous(expression(Metabolic~rate~(HJ/day)))+ scale_x_discrete('')+ theme_classic()+theme(axis.title.y=element_text(vjust=2, size=rel(1.25), face='bold'))

When the distributional assumptions are violated, parametric tests are unreliable. Under these circumstances, non-parametric tests can be very useful.
