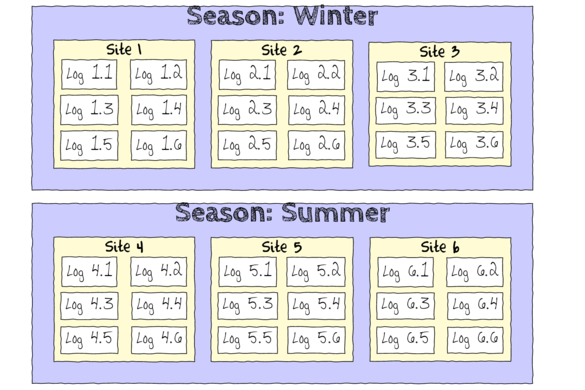
Nested design
Simple
Nested
Simple

Nested

> data.nest <- read.csv('../data/data.nest.csv')
> head(data.nest) y Region Sites Quads
1 32.25789 A S1 1
2 32.40160 A S1 2
3 37.20174 A S1 3
4 36.58866 A S1 4
5 35.45206 A S1 5
6 37.07744 A S1 6> library(ggplot2)
> data.nest$Sites <- factor(data.nest$Sites, levels=paste0('S',1:nSites))
> ggplot(data.nest, aes(y=y, x=1,color=Region)) + geom_boxplot() +
+ facet_grid(.~Sites) +
+ scale_x_continuous('', breaks=NULL)+theme_classic()
> #Effects of treatment
> library(plyr)
> boxplot(y~Region, ddply(data.nest, ~Region+Sites,
+ numcolwise(mean, na.rm=T)))
> #Site effects
> boxplot(y~Sites, ddply(data.nest, ~Region+Sites+Quads,
+ numcolwise(mean, na.rm=T))) 

\(y = \mu + \alpha + \beta(\alpha) + \epsilon\)
e.g.
\(\mu\) - base (mean of first Region)
\(\alpha\) - main fixed effect
\(\beta\) - sub-replicates (Sites: random effect)
> with(data.nest, table(Region,Sites)) Sites
Region S1 S2 S3 S4 S5 S6 S7 S8 S9 S10 S11 S12 S13 S14 S15
A 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0
B 0 0 0 0 0 10 10 10 10 10 0 0 0 0 0
C 0 0 0 0 0 0 0 0 0 0 10 10 10 10 10> head(data.nest, 20) y Region Sites Quads
1 32.25789 A S1 1
2 32.40160 A S1 2
3 37.20174 A S1 3
4 36.58866 A S1 4
5 35.45206 A S1 5
6 37.07744 A S1 6
7 36.39324 A S1 7
8 32.85538 A S1 8
9 22.53580 A S1 9
10 35.58168 A S1 10
11 41.92308 A S2 11
12 41.42474 A S2 12
13 34.84996 A S2 13
14 39.81297 A S2 14
15 44.29343 A S2 15
16 48.99712 A S2 16
17 41.68978 A S2 17
18 44.14208 A S2 18
19 41.93469 A S2 19
20 35.31842 A S2 20> library(nlme)
> data.nest.lme <- lme(y~Region, random=~1|Sites, data.nest)
> plot(data.nest.lme)
> plot(data.nest$Region, residuals(data.nest.lme,
+ type='normalized'))
> summary(data.nest.lme)Linear mixed-effects model fit by REML
Data: data.nest
AIC BIC logLik
927.7266 942.6788 -458.8633
Random effects:
Formula: ~1 | Sites
(Intercept) Residual
StdDev: 13.6582 4.372252
Fixed effects: y ~ Region
Value Std.Error DF t-value p-value
(Intercept) 42.27936 6.139350 135 6.886618 0.0000
RegionB 29.84692 8.682352 12 3.437654 0.0049
RegionC 37.02026 8.682352 12 4.263851 0.0011
Correlation:
(Intr) ReginB
RegionB -0.707
RegionC -0.707 0.500
Standardized Within-Group Residuals:
Min Q1 Med Q3 Max
-2.603787242 -0.572951701 0.004953998 0.620914933 2.765601716
Number of Observations: 150
Number of Groups: 15 > VarCorr(data.nest.lme)Sites = pdLogChol(1)
Variance StdDev
(Intercept) 186.54644 13.658200
Residual 19.11659 4.372252> anova(data.nest.lme) numDF denDF F-value p-value
(Intercept) 1 135 331.8308 <.0001
Region 2 12 10.2268 0.0026> library(multcomp)
> summary(glht(data.nest.lme, linfct=mcp(Region="Tukey")))
Simultaneous Tests for General Linear Hypotheses
Multiple Comparisons of Means: Tukey Contrasts
Fit: lme.formula(fixed = y ~ Region, data = data.nest, random = ~1 |
Sites)
Linear Hypotheses:
Estimate Std. Error z value Pr(>|z|)
B - A == 0 29.847 8.682 3.438 0.00172 **
C - A == 0 37.020 8.682 4.264 < 0.001 ***
C - B == 0 7.173 8.682 0.826 0.68674
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- single-step method)> library(effects)
> plot(allEffects(data.nest.lme))
Step 1. gather model coefficients and model matrix
> coefs <- fixef(data.nest.lme)
> coefs(Intercept) RegionB RegionC
42.27936 29.84692 37.02026 > xs <- levels(data.nest$Region)
> Xmat <- model.matrix(~Region, data.frame(Region=xs))
> head(Xmat) (Intercept) RegionB RegionC
1 1 0 0
2 1 1 0
3 1 0 1Step 3. calculate predicted y and CI
> ys <- t(coefs %*% t(Xmat))
> head(ys) [,1]
1 42.27936
2 72.12628
3 79.29961> SE <- sqrt(diag(Xmat %*% vcov(data.nest.lme) %*% t(Xmat)))
> CI <- 2*SE
> #OR
> CI <- qt(0.975,length(data.nest$y)-2)*SE
> data.nest.pred <- data.frame(Region=xs, fit=ys, se=SE,
+ lower=ys-CI, upper=ys+CI)
> head(data.nest.pred) Region fit se lower upper
1 A 42.27936 6.13935 30.14725 54.41147
2 B 72.12628 6.13935 59.99417 84.25839
3 C 79.29961 6.13935 67.16751 91.43172Step 4. plot it
> with(data.nest.pred,plot.default(Region, fit,type='n',axes=F, ann=F,ylim=range(c(data.nest.pred$lower, data.nest.pred$upper))))
> points(y~Region, data=data.nest, pch=16, col='grey')
> points(fit~Region, data=data.nest.pred, pch=16)
> with(data.nest.pred, arrows(as.numeric(Region),lower,as.numeric(Region),upper, length=0.1, angle=90, code=3))
> axis(1, at=1:3, labels=levels(data.nest$Region))
> mtext('Region',1,line=3)
> axis(2,las=1)
> mtext('Y',2,line=3)
> box(bty='l')

Step 4. plot it
> data.nest$res <- predict(data.nest.lme, level=0)-
+ residuals(data.nest.lme)
> with(data.nest.pred,plot.default(Region, fit,type='n',axes=F, ann=F,ylim=range(c(data.nest.pred$lower, data.nest.pred$upper))))
> points(res~Region, data=data.nest, pch=16, col='grey')
> points(fit~Region, data=data.nest.pred, pch=16)
> with(data.nest.pred, arrows(as.numeric(Region),lower,as.numeric(Region),upper, length=0.1, angle=90, code=3))
> axis(1, at=1:3, labels=levels(data.nest$Region))
> mtext('Region',1,line=3)
> axis(2,las=1)
> mtext('Y',2,line=3)
> box(bty='l')

> library(ggplot2)
> data.nest$res <- predict(data.nest.lme, level=0)-
+ residuals(data.nest.lme)
>
> ggplot(data.nest.pred, aes(y=fit, x=Region))+
+ geom_point(data=data.nest, aes(y=res), col='grey',position = position_jitter(height=0))+
+ geom_errorbar(aes(ymin=lower, ymax=upper), width=0.1)+
+ geom_point()+
+ theme_classic()+
+ theme(axis.title.y=element_text(vjust=2),
+ axis.title.x=element_text(vjust=-1))

| Format of curdies.csv data files | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|

|
||||||||||||||||||||||||||||||||||||||||||||